Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes
- PMID: 34580445
- PMCID: PMC8993687
- DOI: 10.1038/s41564-021-00966-0
Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes
Erratum in
-
Author Correction: Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes.Nat Microbiol. 2022 May;7(5):736. doi: 10.1038/s41564-022-01118-8. Nat Microbiol. 2022. PMID: 35388189 Free PMC article. No abstract available.
Abstract
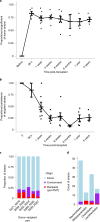
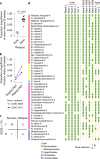
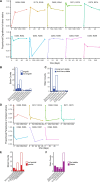
Fecal microbiota transplantation (FMT) has been successfully applied to treat recurrent Clostridium difficile infection in humans, but a precise method to measure which bacterial strains stably engraft in recipients and evaluate their association with clinical outcomes is lacking. We assembled a collection of >1,000 different bacterial strains that were cultured from the fecal samples of 22 FMT donors and recipients. Using our strain collection combined with metagenomic sequencing data from the same samples, we developed a statistical approach named Strainer for the detection and tracking of bacterial strains from metagenomic sequencing data. We applied Strainer to evaluate a cohort of 13 FMT longitudinal clinical interventions and detected stable engraftment of 71% of donor microbiota strains in recipients up to 5 years post-FMT. We found that 80% of recipient gut bacterial strains pre-FMT were eliminated by FMT and that post-FMT the strains present persisted up to 5 years later, together with environmentally acquired strains. Quantification of donor bacterial strain engraftment in recipients independently explained (precision 100%, recall 95%) the clinical outcomes (relapse or success) after initial and repeat FMT. We report a compendium of bacterial species and strains that consistently engraft in recipients over time that could be used in defined live biotherapeutic products as an alternative to FMT. Our analytical framework and Strainer can be applied to systematically evaluate either FMT or defined live bacterial therapeutic studies by quantification of strain engraftment in recipients.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
J.F. is on the scientific advisory board of Vedanta Biosciences, reports receiving research grants from Janssen Pharmaceuticals and reports receiving consulting fees from Innovation Pharmaceuticals, Janssen Pharmaceuticals, BiomX and Vedanta Biosciences. J.-F.C. reports receiving research grants from AbbVie, Janssen Pharmaceuticals and Takeda, receiving payment for lectures from AbbVie, Amgen, Allergan, Bristol Myers Squibb, Ferring Pharmaceuticals, Shire and Takeda, receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Celltrion Healthcare, Eli Lilly, Enterome, Ferring Pharmaceuticals, Geneva Pharmaceuticals, Genentech, Gilead, Iterative Scopes, Ipsen, Imedex, Immunic, Inotrem, Janssen Pharmaceuticals, Landos, LimmaTech Biologics AG, Medimmune, Merck, Novartis, O Mass, Otsuka Pharmaceutical, Pfizer, Shire, Takeda, TiGenix and Viela Bio and holds stock options in Intestinal Biotech Development. D.G. is an employee of Janssen Research and Development. The other authors declare no competing interests. A patent has been filed on this work (Patent Cooperation Treaty application PCT/US21/71018, filed 27 July 2021).
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

