Diverse wMel variants of Wolbachia pipientis differentially rescue fertility and cytological defects of the bag of marbles partial loss of function mutation in Drosophila melanogaster
- PMID: 34580706
- PMCID: PMC8664471
- DOI: 10.1093/g3journal/jkab312
Diverse wMel variants of Wolbachia pipientis differentially rescue fertility and cytological defects of the bag of marbles partial loss of function mutation in Drosophila melanogaster
Abstract
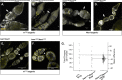
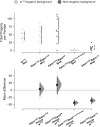
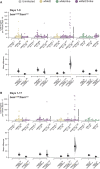
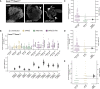
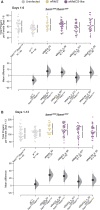
In Drosophila melanogaster, the maternally inherited endosymbiont Wolbachia pipientis interacts with germline stem cell genes during oogenesis. One such gene, bag of marbles (bam) is the key switch for differentiation and also shows signals of adaptive evolution for protein diversification. These observations have led us to hypothesize that W. pipientis could be driving the adaptive evolution of bam for control of oogenesis. To test this hypothesis, we must understand the specificity of the genetic interaction between bam and W. pipientis. Previously, we documented that the W. pipientis variant, wMel, rescued the fertility of the bamBW hypomorphic mutant as a transheterozygote over a bam null. However, bamBW was generated more than 20 years ago in an uncontrolled genetic background and maintained over a balancer chromosome. Consequently, the chromosome carrying bamBW accumulated mutations that have prevented controlled experiments to further assess the interaction. Here, we used CRISPR/Cas9 to engineer the same single amino acid bam hypomorphic mutation (bamL255F) and a new bam null disruption mutation into the w1118 isogenic background. We assess the fertility of wildtype bam, bamL255F/bamnull hypomorphic, and bamL255F/bamL255F mutant females, each infected individually with 10 W. pipientis wMel variants representing three phylogenetic clades. Overall, we find that all of the W. pipientis variants tested here rescue bam hypomorphic fertility defects with wMelCS-like variants exhibiting the strongest rescue effects. In addition, these variants did not increase wildtype bam female fertility. Therefore, both bam and W. pipientis interact in genotype-specific ways to modulate female fertility, a critical fitness phenotype.
Keywords: Wolbachia; bam; differentiation; germline stem cell; oogenesis.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


References
-
- Arnold PA, Levin SC, Stevanovic AL, Johnson KN.. 2019. Drosophila melanogaster infected with Wolbachia strain wMelCS prefer cooler temperatures. Ecol Entomol. 44:287–290. doi:10.1111/een.12696.
-
- Bailly-Bechet M, Martins-Simões P, Szöllosi GJ, Mialdea G, Sagot MF, et al.2017. How long does Wolbachia remain on board? Mol Biol Evol. 34:1183–1193. doi:10.1093/molbev/msx073. - PubMed
-
- Bauer DuMont VL, Flores HA, Wright MH, Aquadro CF.. 2007. Recurrent positive selection at bgcn, a key determinant of germ line differentiation, does not appear to be driven by simple coevolution with its partner protein Bam. Mol Biol Evol. 24:182–191. doi:10.1093/molbev/msl141. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
