Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic
- PMID: 34580920
- PMCID: PMC8661935
- DOI: 10.1002/prot.26250
Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic
Abstract
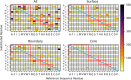
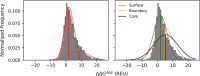
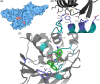
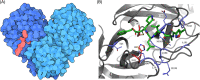
Understanding the molecular evolution of the SARS-CoV-2 virus as it continues to spread in communities around the globe is important for mitigation and future pandemic preparedness. Three-dimensional structures of SARS-CoV-2 proteins and those of other coronavirusess archived in the Protein Data Bank were used to analyze viral proteome evolution during the first 6 months of the COVID-19 pandemic. Analyses of spatial locations, chemical properties, and structural and energetic impacts of the observed amino acid changes in >48 000 viral isolates revealed how each one of 29 viral proteins have undergone amino acid changes. Catalytic residues in active sites and binding residues in protein-protein interfaces showed modest, but significant, numbers of substitutions, highlighting the mutational robustness of the viral proteome. Energetics calculations showed that the impact of substitutions on the thermodynamic stability of the proteome follows a universal bi-Gaussian distribution. Detailed results are presented for potential drug discovery targets and the four structural proteins that comprise the virion, highlighting substitutions with the potential to impact protein structure, enzyme activity, and protein-protein and protein-nucleic acid interfaces. Characterizing the evolution of the virus in three dimensions provides testable insights into viral protein function and should aid in structure-based drug discovery efforts as well as the prospective identification of amino acid substitutions with potential for drug resistance.
Keywords: COVID-19; SARS-CoV-2; coronavirus; databases; evolution; molecular; pandemics; protein; viral proteins.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Update of
-
Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first six months of the COVID-19 pandemic.bioRxiv [Preprint]. 2020 Dec 7:2020.12.01.406637. doi: 10.1101/2020.12.01.406637. bioRxiv. 2020. Update in: Proteins. 2022 May;90(5):1054-1080. doi: 10.1002/prot.26250. PMID: 33299989 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

