Enhanced copper-resistance gene repertoire in Alteromonas macleodii strains isolated from copper-treated marine coatings
- PMID: 34582496
- PMCID: PMC8478169
- DOI: 10.1371/journal.pone.0257800
Enhanced copper-resistance gene repertoire in Alteromonas macleodii strains isolated from copper-treated marine coatings
Abstract
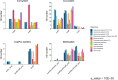
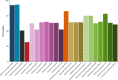
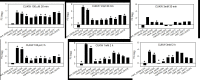
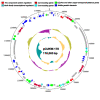
Copper is prevalent in coastal ecosystems due to its use as an algaecide and as an anti-fouling agent on ship hulls. Alteromonas spp. have previously been shown to be some of the early colonizers of copper-based anti-fouling paint but little is known about the mechanisms they use to overcome this initial copper challenge. The main models of copper resistance include the Escherichia coli chromosome-based Cue and Cus systems; the plasmid-based E. coli Pco system; and the plasmid-based Pseudomonas syringae Cop system. These were all elucidated from strains isolated from copper-rich environments of agricultural and/or enteric origin. In this work, copper resistance assays demonstrated the ability of Alteromonas macleodii strains CUKW and KCC02 to grow at levels lethal to other marine bacterial species. A custom database of Hidden Markov Models was designed based on proteins from the Cue, Cus, and Cop/Pco systems and used to identify potential copper resistance genes in CUKW and KCC02. Comparative genomic analyses with marine bacterial species and bacterial species isolated from copper-rich environments demonstrated that CUKW and KCC02 possess genetic elements of all systems, oftentimes with multiple copies, distributed throughout the chromosome and mega-plasmids. In particular, two copies of copA (the key player in cytoplasmic detoxification), each with its own apparent MerR-like transcriptional regulator, occur on a mega-plasmid, along with multiple copies of Pco homologs. Genes from both systems were induced upon exposure to elevated copper levels (100 μM- 3 mM). Genomic analysis identified one of the merR-copA clusters occurs on a genomic island (GI) within the plasmid, and comparative genomic analysis found that either of the merR-copA clusters, which also includes genes coding for a cupredoxin domain-containing protein and an isoprenylcysteine methyltransferase, occurs on a GI across diverse bacterial species. These genomic findings combined with the ability of CUKW and KCC02 to grow in copper-challenged conditions are couched within the context of the genome flexibility of the Alteromonas genus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

