Genome organization and evolution of a eukaryotic nicotinate co-inducible pathway
- PMID: 34582709
- PMCID: PMC8478523
- DOI: 10.1098/rsob.210099
Genome organization and evolution of a eukaryotic nicotinate co-inducible pathway
Abstract
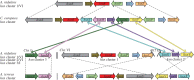
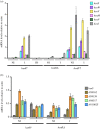
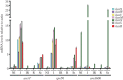
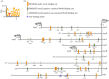
In Aspergillus nidulans a regulon including 11 hxn genes (hxnS, T, R, P, Y, Z, X, W, V, M and N) is inducible by a nicotinate metabolic derivative, repressible by ammonium and under stringent control of the nitrogen-state-sensitive GATA factor AreA and the specific transcription factor HxnR. This is the first report in a eukaryote of the genomic organization of a possibly complete pathway of nicotinate utilization. In A. nidulans the regulon is organized in three distinct clusters, this organization is variable in the Ascomycota. In some Pezizomycotina species all 11 genes map in a single cluster; in others they map in two clusters. This variable organization sheds light on cluster evolution. Instances of gene duplication followed by or simultaneous with integration in the cluster, partial or total cluster loss, and horizontal gene transfer of several genes (including an example of whole cluster re-acquisition in Aspergillus of section Flavi) were detected, together with the incorporation in some clusters of genes not found in the A. nidulans co-regulated regulon, which underlie both the plasticity and the reticulate character of metabolic cluster evolution. This study provides a comprehensive phylogeny of six members of the cluster across representatives of all Ascomycota classes.
Keywords: Aspergillus nidulans; ascomycetes; eukaryotic nicotinate utilization; gene cluster co-regulation; gene cluster evolution; horizontal gene transmission.
Conflict of interest statement
We have no competing interests.
Figures


Similar articles
-
A complete nicotinate degradation pathway in the microbial eukaryote Aspergillus nidulans.Commun Biol. 2022 Jul 21;5(1):723. doi: 10.1038/s42003-022-03684-3. Commun Biol. 2022. PMID: 35864155 Free PMC article.
-
A eukaryotic nicotinate-inducible gene cluster: convergent evolution in fungi and bacteria.Open Biol. 2017 Dec;7(12):170199. doi: 10.1098/rsob.170199. Open Biol. 2017. PMID: 29212709 Free PMC article.
-
The hxB gene, necessary for the post-translational activation of purine hydroxylases in Aspergillus nidulans, is independently controlled by the purine utilization and the nicotinate utilization transcriptional activating systems.Mol Microbiol. 1999 Feb;31(4):1065-73. doi: 10.1046/j.1365-2958.1999.01242.x. Mol Microbiol. 1999. PMID: 10096075
-
Presence, Mode of Action, and Application of Pathway Specific Transcription Factors in Aspergillus Biosynthetic Gene Clusters.Int J Mol Sci. 2021 Aug 13;22(16):8709. doi: 10.3390/ijms22168709. Int J Mol Sci. 2021. PMID: 34445420 Free PMC article. Review.
-
Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation.Prog Nucleic Acid Res Mol Biol. 2001;69:149-204. doi: 10.1016/s0079-6603(01)69047-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550794 Review.
Cited by
-
A complete nicotinate degradation pathway in the microbial eukaryote Aspergillus nidulans.Commun Biol. 2022 Jul 21;5(1):723. doi: 10.1038/s42003-022-03684-3. Commun Biol. 2022. PMID: 35864155 Free PMC article.
-
A non-canonical fungal peroxisome PTS-1 signal, SYM, and its evolutionary aspects.Sci Rep. 2025 Aug 1;15(1):28088. doi: 10.1038/s41598-025-13871-x. Sci Rep. 2025. PMID: 40750654 Free PMC article.
-
Generation, Transfer, and Loss of Alternative Oxidase Paralogues in the Aspergillaceae Family.J Fungi (Basel). 2023 Dec 14;9(12):1195. doi: 10.3390/jof9121195. J Fungi (Basel). 2023. PMID: 38132795 Free PMC article.
-
Regulation of nutrient utilization in filamentous fungi.Appl Microbiol Biotechnol. 2023 Oct;107(19):5873-5898. doi: 10.1007/s00253-023-12680-4. Epub 2023 Aug 4. Appl Microbiol Biotechnol. 2023. PMID: 37540250 Free PMC article. Review.
References
-
- Jimenez JI, Canales A, Jimenez-Barbero J, Ginalski K, Rychlewski L, Garcia JL, Diaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc. Natl Acad. Sci. USA 105, 11 329-11 334. (10.1073/pnas.0802273105) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

