Genome-wide Association Study Identifies 2 New Loci Associated With Anti-NMDAR Encephalitis
- PMID: 34584012
- PMCID: PMC8479862
- DOI: 10.1212/NXI.0000000000001085
Genome-wide Association Study Identifies 2 New Loci Associated With Anti-NMDAR Encephalitis
Abstract
Background and objectives: To investigate the genetic determinants of the most common type of antibody-mediated autoimmune encephalitis, anti-NMDA receptor (anti-NMDAR) encephalitis.
Methods: We performed a genome-wide association study in 178 patients with anti-NMDAR encephalitis and 590 healthy controls, followed by a colocalization analysis to identify putatively causal genes.
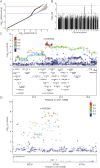
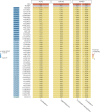
Results: We identified 2 independent risk loci harboring genome-wide significant variants (p < 5 × 10-8, OR ≥ 2.2), 1 on chromosome 15, harboring only the LRRK1 gene, and 1 on chromosome 11 centered on the ACP2 and NR1H3 genes in a larger region of high linkage disequilibrium. Colocalization signals with expression quantitative trait loci for different brain regions and immune cell types suggested ACP2, NR1H3, MADD, DDB2, and C11orf49 as putatively causal genes. The best candidate genes in each region are LRRK1, encoding leucine-rich repeat kinase 1, a protein involved in B-cell development, and NR1H3 liver X receptor alpha, a transcription factor whose activation inhibits inflammatory processes.
Discussion: This study provides evidence for relevant genetic determinants of antibody-mediated autoimmune encephalitides outside the human leukocyte antigen (HLA) region. The results suggest that future studies with larger sample sizes will successfully identify additional genetic determinants and contribute to the elucidation of the pathomechanism.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Figures

References
-
- Dalmau J, Armangué T, Planagumà J, et al. An update on anti-NMDA receptor encephalitis for neurologists and psychiatrists: mechanisms and models. Lancet Neurol. 2019;18(11):1045-1057. - PubMed
-
- Mueller SH, Färber A, Prüss H, et al. Genetic predisposition in anti-LGI1 and anti-NMDA receptor encephalitis. Ann Neurol. 2018;83(4):863-869. - PubMed
-
- Nothlings U, Krawczak M, PopGen. A population-based biobank with prospective follow-up of a control group. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55(6-7):831-835. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
