Do second generation sequencing techniques identify documented genetic markers for neonatal diabetes mellitus?
- PMID: 34584998
- PMCID: PMC8455689
- DOI: 10.1016/j.heliyon.2021.e07903
Do second generation sequencing techniques identify documented genetic markers for neonatal diabetes mellitus?
Abstract
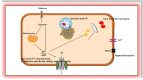
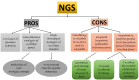
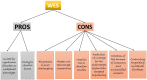
Neonatal diabetes mellitus (NDM) is noted as a genetic, heterogeneous, and rare disease in infants. NDM occurs due to a single-gene mutation in neonates. A common source for developing NDM in an infant is the existence of mutations/variants in the KCNJ11 and ABCC8 genes, encoding the subunits of the voltage-dependent potassium channel. Both KCNJ11 and ABCC8 genes are useful in diagnosing monogenic diabetes during infancy. Genetic analysis was previously performed using first-generation sequencing techniques, such as DNA-Sanger sequencing, which uses chain-terminating inhibitors. Sanger sequencing has certain limitations; it can screen a limited region of exons in one gene, but it cannot screen large regions of the human genome. In the last decade, first generation sequencing techniques have been replaced with second-generation sequencing techniques, such as next-generation sequencing (NGS), which sequences nucleic-acids more rapidly and economically than Sanger sequencing. NGS applications are involved in whole exome sequencing (WES), whole genome sequencing (WGS), and targeted gene panels. WES characterizes a substantial breakthrough in human genetics. Genetic testing for custom genes allows the screening of the complete gene, including introns and exons. The aim of this review was to confirm if the 22 genetic variations previously documented to cause NDM by Sanger sequencing could be detected using second generation sequencing techniques. The author has cross-checked global studies performed in NDM using NGS, ES/WES, WGS, and targeted gene panels as second-generation sequencing techniques; WES confirmed the similar variants, which have been previously documented with Sanger sequencing. WES is documented as a powerful tool and WGS as the most comprehensive test for verified the documented variants, as well as novel enhancers. This review recommends for the future studies should be performed with second generation sequencing techniques to identify the verified 22 genetic and novel variants by screening in NDM (PNDM or TNMD) children.
Keywords: DNA-Sanger sequencing; Exome sequencing (ES); First-generation sequencing; Neonatal diabetes mellitus (NDM); Next-generation sequencing (NGS); Second-generation sequencing; Targeted gene panels; Whole exome sequencing (WES); Whole genome sequencing (WGS).
© 2021 The Author.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Al-Fadhli F.M., Afqi M., Sairafi M.H., Almuntashri M., Alharby E., Alharbi G., Abdud Samad F., Hashmi J.A., Zaytuni D., Bahashwan A.A. Biallelic loss of function variant in the unfolded protein response gene Pdia6 is associated with asphyxiating thoracic dystrophy and neonatal-onset diabetes. Clin. Genet. 2021;99:694–703. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

