Activated PI3Kδ signals compromise plasma cell survival via limiting autophagy and increasing ER stress
- PMID: 34586341
- PMCID: PMC8485856
- DOI: 10.1084/jem.20211035
Activated PI3Kδ signals compromise plasma cell survival via limiting autophagy and increasing ER stress
Abstract
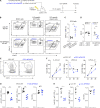
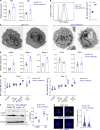
While phosphatidylinositide 3-kinase delta (PI3Kδ) plays a critical role in humoral immunity, the requirement for PI3Kδ signaling in plasma cells remains poorly understood. Here, we used a conditional mouse model of activated PI3Kδ syndrome (APDS), to interrogate the function of PI3Kδ in plasma cell biology. Mice expressing a PIK3CD gain-of-function mutation (aPIK3CD) in B cells generated increased numbers of memory B cells and mounted an enhanced secondary response but exhibited a rapid decay of antibody levels over time. Consistent with these findings, aPIK3CD expression markedly impaired plasma cell generation, and expression of aPIK3CD intrinsically in plasma cells was sufficient to diminish humoral responses. Mechanistically, aPIK3CD disrupted ER proteostasis and autophagy, which led to increased plasma cell death. Notably, this defect was driven primarily by elevated mTORC1 signaling and modulated by treatment with PI3Kδ-specific inhibitors. Our findings establish an essential role for PI3Kδ in plasma cell homeostasis and suggest that modulating PI3Kδ activity may be useful for promoting and/or thwarting specific immune responses.
© 2021 Al Qureshah et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

