G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy
- PMID: 34586841
- PMCID: PMC8480930
- DOI: 10.1126/sciadv.abh3243
G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy
Abstract
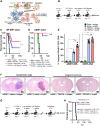
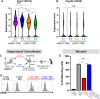
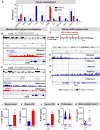
Mutant isocitrate-dehydrogenase 1 (mIDH1) synthesizes the oncometabolite 2-hydroxyglutarate (2HG), which elicits epigenetic reprogramming of the glioma cells’ transcriptome by inhibiting DNA and histone demethylases. We show that the efficacy of immune-stimulatory gene therapy (TK/Flt3L) is enhanced in mIDH1 gliomas, due to the reprogramming of the myeloid cells’ compartment infiltrating the tumor microenvironment (TME). We uncovered that the immature myeloid cells infiltrating the mIDH1 TME are mainly nonsuppressive neutrophils and preneutrophils. Myeloid cell reprogramming was triggered by granulocyte colony-stimulating factor (G-CSF) secreted by mIDH1 glioma stem/progenitor-like cells. Blocking G-CSF in mIDH1 glioma–bearing mice restores the inhibitory potential of the tumor-infiltrating myeloid cells, accelerating tumor progression. We demonstrate that G-CSF reprograms bone marrow granulopoiesis, resulting in noninhibitory myeloid cells within mIDH1 glioma TME and enhancing the efficacy of immune-stimulatory gene therapy.
Figures







References
-
- Ceccarelli M., Barthel F. P., Malta T. M., Sabedot T. S., Salama S. R., Murray B. A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S. M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., Rao A. A., Grifford M., Cherniack A. D., Zhang H., Poisson L., Carlotti C. G. Jr., Tirapelli D. P. C., Rao A., Mikkelsen T., Lau C. C., Yung W. K. A., Rabadan R., Huse J., Brat D. J., Lehman N. L., Barnholtz-Sloan J. S., Zheng S., Hess K., Rao G., Meyerson M., Beroukhim R., Cooper L., Akbani R., Wrensch M., Haussler D., Aldape K. D., Laird P. W., Gutmann D. H., Noushmehr H., Iavarone A., Verhaak R. G. W., Anjum S., Arachchi H., Auman J. T., Balasundaram M., Balu S., Barnett G., Baylin S., Bell S., Benz C., Bir N., Black K. L., Bodenheimer T., Boice L., Bootwalla M. S., Bowen J., Bristow C. A., Butterfield Y. S. N., Chen Q. R., Chin L., Cho J., Chuah E., Chudamani S., Coetzee S. G., Cohen M. L., Colman H., Couce M., D’Angelo F., Davidsen T., Davis A., Demchok J. A., Devine K., Ding L., Duell R., Elder J. B., Eschbacher J. M., Fehrenbach A., Ferguson M., Frazer S., Fuller G., Fulop J., Gabriel S. B., Garofano L., Gastier-Foster J. M., Gehlenborg N., Gerken M., Getz G., Giannini C., Gibson W. J., Hadjipanayis A., Hayes D. N., Heiman D. I., Hermes B., Hilty J., Hoadley K. A., Hoyle A. P., Huang M., Jefferys S. R., Jones C. D., Jones S. J. M., Ju Z., Kastl A., Kendler A., Kim J., Kucherlapati R., Lai P. H., Lawrence M. S., Lee S., Leraas K. M., Lichtenberg T. M., Lin P., Liu Y., Liu J., Ljubimova J. Y., Lu Y., Ma Y., Maglinte D. T., Mahadeshwar H. S., Marra M. A., McGraw M., McPherson C., Meng S., Mieczkowski P. A., Miller C. R., Mills G. B., Moore R. A., Mose L. E., Mungall A. J., Naresh R., Naska T., Neder L., Noble M. S., Noss A., O’Neill B. P., Ostrom Q. T., Palmer C., Pantazi A., Parfenov M., Park P. J., Parker J. S., Perou C. M., Pierson C. R., Pihl T., Protopopov A., Radenbaugh A., Ramirez N. C., Rathmell W. K., Ren X., Roach J., Robertson A. G., Saksena G., Schein J. E., Schumacher S. E., Seidman J., Senecal K., Seth S., Shen H., Shi Y., Shih J., Shimmel K., Sicotte H., Sifri S., Silva T., Simons J. V., Singh R., Skelly T., Sloan A. E., Sofia H. J., Soloway M. G., Song X., Sougnez C., Souza C., Staugaitis S. M., Sun H., Sun C., Tan D., Tang J., Tang Y., Thorne L., Trevisan F. A., Triche T., van den Berg D. J., Veluvolu U., Voet D., Wan Y., Wang Z., Warnick R., Weinstein J. N., Weisenberger D. J., Wilkerson M. D., Williams F., Wise L., Wolinsky Y., Wu J., Xu A. W., Yang L., Yang L., Zack T. I., Zenklusen J. C., Zhang J., Zhang W., Zhang J., Zmuda E., Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016). - PMC - PubMed
-
- Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B., Bigner D. D., IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009). - PMC - PubMed
-
- Figueroa M. E., Abdel-Wahab O., Lu C., Ward P. S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H. F., Tallman M. S., Sun Z., Wolniak K., Peeters J. K., Liu W., Choe S. E., Fantin V. R., Paietta E., Löwenberg B., Licht J. D., Godley L. A., Delwel R., Valk P. J. M., Thompson C. B., Levine R. L., Melnick A., Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010). - PMC - PubMed
-
- Lu C., Ward P. S., Kapoor G. S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C. R., Khanin R., Figueroa M. E., Melnick A., Wellen K. E., O’Rourke D. M., Berger S. L., Chan T. A., Levine R. L., Mellinghoff I. K., Thompson C. B., IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012). - PMC - PubMed
-
- Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., Thompson C. B., The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010). - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

