Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus
- PMID: 34586857
- PMCID: PMC8754229
- DOI: 10.1128/JVI.01253-21
Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus
Abstract
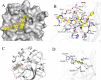
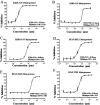
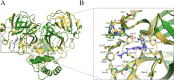
Over the past 20 years, the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2 emerged, causing severe human respiratory diseases throughout the globe. Developing broad-spectrum drugs would be invaluable in responding to new, emerging coronaviruses and to address unmet urgent clinical needs. Main protease (Mpro; also known as 3CLpro) has a major role in the coronavirus life cycle and is one of the most important targets for anti-coronavirus agents. We show that a natural product, noncovalent inhibitor, shikonin, is a pan-main protease inhibitor of SARS-CoV-2, SARS-CoV, MERS-CoV, human coronavirus (HCoV)-HKU1, HCoV-NL63, and HCoV-229E with micromolar half maximal inhibitory concentration (IC50) values. Structures of the main protease of different coronavirus genus, SARS-CoV from the betacoronavirus genus and HCoV-NL63 from the alphacoronavirus genus, were determined by X-ray crystallography and revealed that the inhibitor interacts with key active site residues in a unique mode. The structure of the main protease inhibitor complex presents an opportunity to discover a novel series of broad-spectrum inhibitors. These data provide substantial evidence that shikonin and its derivatives may be effective against most coronaviruses as well as emerging coronaviruses of the future. Given the importance of the main protease for coronavirus therapeutic indication, insights from these studies should accelerate the development and design of safer and more effective antiviral agents. IMPORTANCE The current pandemic has created an urgent need for broad-spectrum inhibitors of SARS-CoV-2. The main protease is relatively conservative compared to the spike protein and, thus, is one of the most promising targets in developing anti-coronavirus agents. We solved the crystal structures of the main protease of SARS-CoV and HCoV-NL63 that bound to shikonin. The structures provide important insights, have broad implications for understanding the structural basis underlying enzyme activity, and can facilitate rational design of broad-spectrum anti-coronavirus ligands as new therapeutic agents.
Keywords: broad-spectrum; coronavirus; inhibitor; main protease; nature product.
Figures


References
-
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. 10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 382:1199–1207. 10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

