Simultaneous and Staggered Foot-and-Mouth Disease Virus Coinfection of Cattle
- PMID: 34586864
- PMCID: PMC8610595
- DOI: 10.1128/JVI.01650-21
Simultaneous and Staggered Foot-and-Mouth Disease Virus Coinfection of Cattle
Abstract
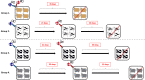
Foot-and-mouth disease (FMD) field studies have suggested the occurrence of simultaneous infection of individual hosts by multiple virus strains; however, the pathogenesis of foot-and-mouth disease virus (FMDV) coinfections is largely unknown. In the current study, cattle were experimentally exposed to two FMDV strains of different serotypes (O and A). One cohort was simultaneously infected with both viruses, while additional cohorts were initially infected with FMDV A and subsequently superinfected with FMDV O after 21 or 35 days. Coinfections were confirmed during acute infection, with both viruses concurrently detected in blood, lesions, and secretions. Staggered exposures resulted in overlapping infections as convalescent animals with persistent subclinical FMDV infection were superinfected with a heterologous virus. Staggering virus exposure by 21 days conferred clinical protection in six of eight cattle, which were subclinically infected following the heterologous virus exposure. This effect was transient, as all animals superinfected at 35 days post-initial infection developed fulminant FMD. The majority of cattle maintained persistent infection with one of the two viruses while clearing the other. Analysis of viral genomes confirmed interserotypic recombination events within 10 days in the upper respiratory tract of five superinfected animals from which the dominant genomes contained the capsid coding regions of the O virus and nonstructural coding regions of the A virus. In contrast, there were no dominant recombinant genomes detected in samples from simultaneously coinfected cattle. These findings inculpate persistently infected carriers as potential FMDV mixing vessels in which novel strains may rapidly emerge through superinfection and recombination. IMPORTANCE Foot-and-mouth disease (FMD) is a viral infection of livestock of critical socioeconomic importance. Field studies from areas of endemic FMD suggest that animals can be simultaneously infected by more than one distinct variant of FMD virus (FMDV), potentially resulting in emergence of novel viral strains through recombination. However, there has been limited investigation of the mechanisms of in vivo FMDV coinfections under controlled experimental conditions. Our findings confirmed that cattle could be simultaneously infected by two distinct serotypes of FMDV, with different outcomes associated with the timing of exposure to the two different viruses. Additionally, dominant interserotypic recombinant FMDVs were discovered in multiple samples from the upper respiratory tracts of five superinfected animals, emphasizing the potential importance of persistently infected FMDV carriers as sources of novel FMDV strains.
Keywords: FMD; FMDV; cattle; coinfection; foot-and-mouth disease; foot-and-mouth disease virus; pathogenesis; persistence; virus.
Figures





References
-
- Arzt J, Baxt B, Grubman MJ, Jackson T, Juleff N, Rhyan J, Rieder E, Waters R, Rodriguez LL. 2011. The pathogenesis of foot-and-mouth disease. II. Viral pathways in swine, small ruminants, and wildlife; myotropism, chronic syndromes, and molecular virus-host interactions. Transbound Emerg Dis 58:305–326. 10.1111/j.1865-1682.2011.01236.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

