Dyrk1a gene dosage in glutamatergic neurons has key effects in cognitive deficits observed in mouse models of MRD7 and Down syndrome
- PMID: 34587162
- PMCID: PMC8480849
- DOI: 10.1371/journal.pgen.1009777
Dyrk1a gene dosage in glutamatergic neurons has key effects in cognitive deficits observed in mouse models of MRD7 and Down syndrome
Abstract
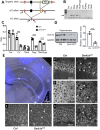
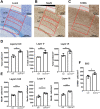
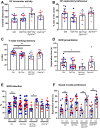
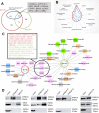
Perturbation of the excitation/inhibition (E/I) balance leads to neurodevelopmental diseases including to autism spectrum disorders, intellectual disability, and epilepsy. Loss-of-function mutations in the DYRK1A gene, located on human chromosome 21 (Hsa21,) lead to an intellectual disability syndrome associated with microcephaly, epilepsy, and autistic troubles. Overexpression of DYRK1A, on the other hand, has been linked with learning and memory defects observed in people with Down syndrome (DS). Dyrk1a is expressed in both glutamatergic and GABAergic neurons, but its impact on each neuronal population has not yet been elucidated. Here we investigated the impact of Dyrk1a gene copy number variation in glutamatergic neurons using a conditional knockout allele of Dyrk1a crossed with the Tg(Camk2-Cre)4Gsc transgenic mouse. We explored this genetic modification in homozygotes, heterozygotes and combined with the Dp(16Lipi-Zbtb21)1Yey trisomic mouse model to unravel the consequence of Dyrk1a dosage from 0 to 3, to understand its role in normal physiology, and in MRD7 and DS. Overall, Dyrk1a dosage in postnatal glutamatergic neurons did not impact locomotor activity, working memory or epileptic susceptibility, but revealed that Dyrk1a is involved in long-term explicit memory. Molecular analyses pointed at a deregulation of transcriptional activity through immediate early genes and a role of DYRK1A at the glutamatergic post-synapse by deregulating and interacting with key post-synaptic proteins implicated in mechanism leading to long-term enhanced synaptic plasticity. Altogether, our work gives important information to understand the action of DYRK1A inhibitors and have a better therapeutic approach.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: SDG is Founder, President and CEO of Proteas Bioanalytics Inc., BioLabs at the Lundquist Institute, 1124 West Carson Street, Torrance, CA, USA.
Figures


References
-
- Ji JL, Lee H, Argiropoulos B, Dorrani N, Mann J, Martinez-Agosto JA, et al.. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. European Journal of Human Genetics. 2015;23(11):1473–81. doi: 10.1038/ejhg.2015.71 WOS:000362916200010. - DOI - PMC - PubMed
-
- Evers JM, Laskowski RA, Bertolli M, Clayton-Smith J, Deshpande C, Eason J, et al.. Structural analysis of pathogenic mutations in the DYRK1A gene in patients with developmental disorders. Hum Mol Genet. 2017;26(3):519–26. doi: 10.1093/hmg/ddw409 ; PubMed Central PMCID: PMC5409128. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

