Development of a fluorogenic ADAMTS-7 substrate
- PMID: 34587841
- PMCID: PMC8494430
- DOI: 10.1080/14756366.2021.1983808
Development of a fluorogenic ADAMTS-7 substrate
Abstract
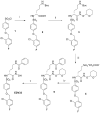
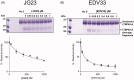
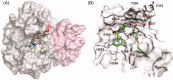
The extracellular protease ADAMTS-7 has been identified as a potential therapeutic target in atherosclerosis and associated diseases such as coronary artery disease (CAD). However, ADAMTS-7 inhibitors have not been reported so far. Screening of inhibitors has been hindered by the lack of a suitable peptide substrate and, consequently, a convenient activity assay. Here we describe the first fluorescence resonance energy transfer (FRET) substrate for ADAMTS-7, ATS7FP7. ATS7FP7 was used to measure inhibition constants for the endogenous ADAMTS-7 inhibitor, TIMP-4, as well as two hydroxamate-based zinc chelating inhibitors. These inhibition constants match well with IC50 values obtained with our SDS-PAGE assay that uses the N-terminal fragment of latent TGF-β-binding protein 4 (LTBP4S-A) as a substrate. Our novel fluorogenic substrate ATS7FP7 is suitable for high throughput screening of ADAMTS-7 inhibitors, thus accelerating translational studies aiming at inhibition of ADAMTS-7 as a novel treatment for cardiovascular diseases such as atherosclerosis and CAD.
Keywords: ADAMTS-7; ADAMTS7; activity assay; coronary artery disease; inhibitor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
