Brief segments of neurophysiological activity enable individual differentiation
- PMID: 34588439
- PMCID: PMC8481307
- DOI: 10.1038/s41467-021-25895-8
Brief segments of neurophysiological activity enable individual differentiation
Abstract
Large, openly available datasets and current analytic tools promise the emergence of population neuroscience. The considerable diversity in personality traits and behaviour between individuals is reflected in the statistical variability of neural data collected in such repositories. Recent studies with functional magnetic resonance imaging (fMRI) have concluded that patterns of resting-state functional connectivity can both successfully distinguish individual participants within a cohort and predict some individual traits, yielding the notion of an individual's neural fingerprint. Here, we aim to clarify the neurophysiological foundations of individual differentiation from features of the rich and complex dynamics of resting-state brain activity using magnetoencephalography (MEG) in 158 participants. We show that akin to fMRI approaches, neurophysiological functional connectomes enable the differentiation of individuals, with rates similar to those seen with fMRI. We also show that individual differentiation is equally successful from simpler measures of the spatial distribution of neurophysiological spectral signal power. Our data further indicate that differentiation can be achieved from brain recordings as short as 30 seconds, and that it is robust over time: the neural fingerprint is present in recordings performed weeks after their baseline reference data was collected. This work, thus, extends the notion of a neural or brain fingerprint to fast and large-scale resting-state electrophysiological dynamics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

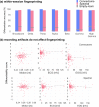
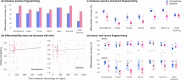
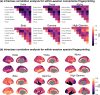

References
-
- Yarkoni, T. in APA handbook of personality and social psychology, Volume 4: Personality Processes and Individual Differences (eds. Mikulincer, M, Shaver, P. R., Cooper, M. L. & Larsen, R. J.) pp. 61–83 (American Psychological Association, Washington, 2015).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

