A random-walk-based epidemiological model
- PMID: 34588487
- PMCID: PMC8481482
- DOI: 10.1038/s41598-021-98211-5
A random-walk-based epidemiological model
Abstract
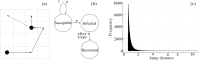
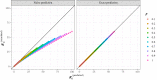
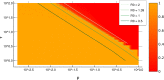
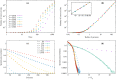
Random walkers on a two-dimensional square lattice are used to explore the spatio-temporal growth of an epidemic. We have found that a simple random-walk system generates non-trivial dynamics compared with traditional well-mixed models. Phase diagrams characterizing the long-term behaviors of the epidemics are calculated numerically. The functional dependence of the basic reproductive number [Formula: see text] on the model's defining parameters reveals the role of spatial fluctuations and leads to a novel expression for [Formula: see text]. Special attention is given to simulations of inter-regional transmission of the contagion. The scaling of the epidemic with respect to space and time scales is studied in detail in the critical region, which is shown to be compatible with the directed-percolation universality class.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Hethcote HW. The mathematics of infectious diseases. SIAM Rev. 2000;42:599–653. doi: 10.1137/S0036144500371907. - DOI
Publication types
LinkOut - more resources
Full Text Sources

