Rhizosphere Soil Bacterial Communities of Continuous Cropping-Tolerant and Sensitive Soybean Genotypes Respond Differently to Long-Term Continuous Cropping in Mollisols
- PMID: 34589076
- PMCID: PMC8473881
- DOI: 10.3389/fmicb.2021.729047
Rhizosphere Soil Bacterial Communities of Continuous Cropping-Tolerant and Sensitive Soybean Genotypes Respond Differently to Long-Term Continuous Cropping in Mollisols
Abstract
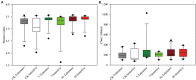
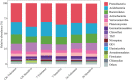
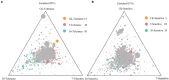
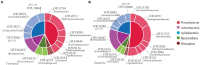
The continuous planting of soybeans leads to soil acidification, aggravation of soil-borne diseases, reduction in soil enzyme activity, and accumulation of toxins in the soil. Microorganisms in the rhizosphere play a very important role in maintaining the sustainability of the soil ecosystem and plant health. In this study, two soybean genotypes, one bred for continuous cropping and the other not, were grown in a Mollisol in northeast China under continuous cropping for 7 and 36years in comparison with soybean-maize rotation, and microbial communities in the rhizosphere composition were assessed using high-throughput sequencing technology. The results showed that short- or long-term continuous cropping had no significant effect on the rhizosphere soil bacterial alpha diversity. Short-term continuous planting increased the number of soybean cyst nematode (Heterodera glycines), while long-term continuous planting reduced these numbers. There were less soybean cyst nematodes in the rhizosphere of the tolerant genotypes than sensitive genotypes. In addition, continuous cropping significantly increased the potential beneficial bacterial populations, such as Pseudoxanthomonas, Nitrospira, and Streptomyces compared to rotation and short-term continuous cropping, suggesting that long-term continuous cropping of soybean shifts the microbial community toward a healthy crop rotation system. Soybean genotypes that are tolerant to soybean might recruit some microorganisms that enhance the resistance of soybeans to long-term continuous cropping. Moreover, the network of the two genotypes responded differently to continuous cropping. The tolerant genotype responded positively to continuous cropping, while for the sensitive genotype, topology analyses on the instability of microbial community in the rhizosphere suggested that short periods of continuous planting can have a detrimental effect on microbial community stability, although this effect could be alleviated with increasing periods of continuous planting.
Keywords: Mollisol; continuous cropping; network; rhizosphere microorganisms; soybean.
Copyright © 2021 Yuan, Yu, Shi, Han, Yu, Wang, Wang, Xiang, Wen, Nian and Lian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The shift of soil microbial community induced by cropping sequence affect soil properties and crop yield.Front Microbiol. 2023 Feb 16;14:1095688. doi: 10.3389/fmicb.2023.1095688. eCollection 2023. Front Microbiol. 2023. PMID: 36910216 Free PMC article.
-
Archaeal communities perform an important role in maintaining microbial stability under long term continuous cropping systems.Sci Total Environ. 2022 Sep 10;838(Pt 3):156413. doi: 10.1016/j.scitotenv.2022.156413. Epub 2022 May 31. Sci Total Environ. 2022. PMID: 35660449
-
Effects of Rhizophagus intraradices on soybean yield and the composition of microbial communities in the rhizosphere soil of continuous cropping soybean.Sci Rep. 2022 Oct 17;12(1):17390. doi: 10.1038/s41598-022-22473-w. Sci Rep. 2022. PMID: 36253456 Free PMC article.
-
Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years.Front Microbiol. 2023 Sep 7;14:1203796. doi: 10.3389/fmicb.2023.1203796. eCollection 2023. Front Microbiol. 2023. PMID: 37744902 Free PMC article. Review.
-
Research progress of rhizosphere microorganisms in Fritillaria L. medicinal plants.Front Bioeng Biotechnol. 2022 Nov 7;10:1054757. doi: 10.3389/fbioe.2022.1054757. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36420438 Free PMC article. Review.
Cited by
-
Land use differentially affects fungal communities and network complexity in northeast China.Front Microbiol. 2022 Nov 18;13:1064363. doi: 10.3389/fmicb.2022.1064363. eCollection 2022. Front Microbiol. 2022. PMID: 36466694 Free PMC article.
-
Maize-peanut rotational strip intercropping improves peanut growth and soil properties by optimizing microbial community diversity.PeerJ. 2022 Jul 28;10:e13777. doi: 10.7717/peerj.13777. eCollection 2022. PeerJ. 2022. PMID: 35919403 Free PMC article.
-
New insights into the occurrence of continuous cropping obstacles in pea (Pisum sativum L.) from soil bacterial communities, root metabolism and gene transcription.BMC Plant Biol. 2023 Apr 28;23(1):226. doi: 10.1186/s12870-023-04225-8. BMC Plant Biol. 2023. PMID: 37106450 Free PMC article.
-
The response of sugar beet rhizosphere micro-ecological environment to continuous cropping.Front Microbiol. 2022 Sep 7;13:956785. doi: 10.3389/fmicb.2022.956785. eCollection 2022. Front Microbiol. 2022. PMID: 36160206 Free PMC article.
-
The shift of soil microbial community induced by cropping sequence affect soil properties and crop yield.Front Microbiol. 2023 Feb 16;14:1095688. doi: 10.3389/fmicb.2023.1095688. eCollection 2023. Front Microbiol. 2023. PMID: 36910216 Free PMC article.
References
-
- Avidano L., Gamalero E., Cossa G. P., Carraro E. (2005). Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30, 21–33. doi: 10.1016/j.apsoil.2005.01.003 - DOI
-
- Cai B. Y., Wang L. Y., Hu W., Jie W. G., Ling H. Z. (2015). Analysis of community structure of root rot pathogenic fungi in seedling stage of soybean continuous cropping. Chin. Agr. Sci. Bull. 31, 92–98.
LinkOut - more resources
Full Text Sources

