A protein interaction landscape of breast cancer
- PMID: 34591612
- PMCID: PMC9040556
- DOI: 10.1126/science.abf3066
A protein interaction landscape of breast cancer
Abstract
Cancers have been associated with a diverse array of genomic alterations. To help mechanistically understand such alterations in breast-invasive carcinoma, we applied affinity purification–mass spectrometry to delineate comprehensive biophysical interaction networks for 40 frequently altered breast cancer (BC) proteins, with and without relevant mutations, across three human breast cell lines. These networks identify cancer-specific protein-protein interactions (PPIs), interconnected and enriched for common and rare cancer mutations, that are substantially rewired by the introduction of key BC mutations. Our analysis identified BPIFA1 and SCGB2A1 as PIK3CA-interacting proteins, which repress PI3K-AKT signaling, and uncovered USP28 and UBE2N as functionally relevant interactors of BRCA1. We also show that the protein phosphatase 1 regulatory subunit spinophilin interacts with and regulates dephosphorylation of BRCA1 to promote DNA double-strand break repair. Thus, PPI landscapes provide a powerful framework for mechanistically interpreting disease genomic data and can identify valuable therapeutic targets.
Figures

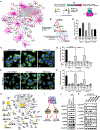

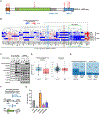

Comment in
-
Identifying cancer drivers.Science. 2021 Oct;374(6563):38-39. doi: 10.1126/science.abl9080. Epub 2021 Sep 30. Science. 2021. PMID: 34591644
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics, 2021. CA: A Cancer Journal for Clinicians. 71 (2021), pp. 7–33. - PubMed
-
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer, Consortium, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ‘t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, The landscape of cancer genes and mutational processes in breast cancer. Nature. 486, 400–404 (2012). - PMC - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JKV, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PVK, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B, The genomic landscapes of human breast and colorectal cancers. Science. 318, 1108–1113 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

