Signal-based optical map alignment
- PMID: 34591846
- PMCID: PMC8483326
- DOI: 10.1371/journal.pone.0253102
Signal-based optical map alignment
Abstract
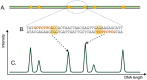
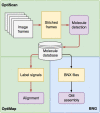
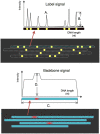
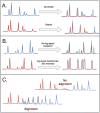
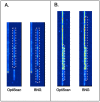
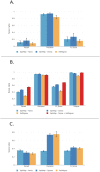
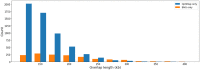
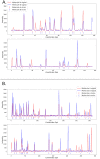
In genomics, optical mapping technology provides long-range contiguity information to improve genome sequence assemblies and detect structural variation. Originally a laborious manual process, Bionano Genomics platforms now offer high-throughput, automated optical mapping based on chips packed with nanochannels through which unwound DNA is guided and the fluorescent DNA backbone and specific restriction sites are recorded. Although the raw image data obtained is of high quality, the processing and assembly software accompanying the platforms is closed source and does not seem to make full use of data, labeling approximately half of the measured signals as unusable. Here we introduce two new software tools, independent of Bionano Genomics software, to extract and process molecules from raw images (OptiScan) and to perform molecule-to-molecule and molecule-to-reference alignments using a novel signal-based approach (OptiMap). We demonstrate that the molecules detected by OptiScan can yield better assemblies, and that the approach taken by OptiMap results in higher use of molecules from the raw data. These tools lay the foundation for a suite of open-source methods to process and analyze high-throughput optical mapping data. The Python implementations of the OptiTools are publicly available through http://www.bif.wur.nl/.
Conflict of interest statement
No authors have competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

