Systematic investigation of cytokine signaling activity at the tissue and single-cell levels
- PMID: 34594031
- PMCID: PMC8493809
- DOI: 10.1038/s41592-021-01274-5
Systematic investigation of cytokine signaling activity at the tissue and single-cell levels
Abstract
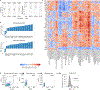
Cytokines are critical for intercellular communication in human health and disease, but the investigation of cytokine signaling activity has remained challenging due to the short half-lives of cytokines and the complexity/redundancy of cytokine functions. To address these challenges, we developed the Cytokine Signaling Analyzer (CytoSig; https://cytosig.ccr.cancer.gov/ ), providing both a database of target genes modulated by cytokines and a predictive model of cytokine signaling cascades from transcriptomic profiles. We collected 20,591 transcriptome profiles for human cytokine, chemokine and growth factor responses. This atlas of transcriptional patterns induced by cytokines enabled the reliable prediction of signaling activities in distinct cell populations in infectious diseases, chronic inflammation and cancer using bulk and single-cell transcriptomic data. CytoSig revealed previously unidentified roles of many cytokines, such as BMP6 as an anti-inflammatory factor, and identified candidate therapeutic targets in human inflammatory diseases, such as CXCL8 for severe coronavirus disease 2019.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Figures

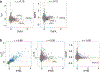


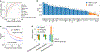
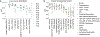






References
-
- Ozaki K & Leonard WJ Cytokine and cytokine receptor pleiotropy and redundancy. J. Biol. Chem 277, 29355–29358 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA163222/CA/NCI NIH HHS/United States
- ZIA BC011889/ImNIH/Intramural NIH HHS/United States
- K12HL138037/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- K12 HL138037/HL/NHLBI NIH HHS/United States
- Z99 CA999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

