High-resolution structural and functional deep brain imaging using adaptive optics three-photon microscopy
- PMID: 34594033
- PMCID: PMC8490155
- DOI: 10.1038/s41592-021-01257-6
High-resolution structural and functional deep brain imaging using adaptive optics three-photon microscopy
Abstract
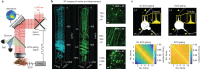
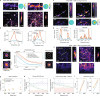
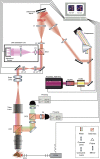
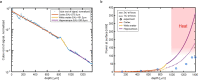
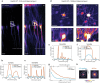
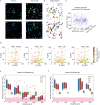
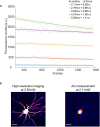
Multiphoton microscopy has become a powerful tool with which to visualize the morphology and function of neural cells and circuits in the intact mammalian brain. However, tissue scattering, optical aberrations and motion artifacts degrade the imaging performance at depth. Here we describe a minimally invasive intravital imaging methodology based on three-photon excitation, indirect adaptive optics (AO) and active electrocardiogram gating to advance deep-tissue imaging. Our modal-based, sensorless AO approach is robust to low signal-to-noise ratios as commonly encountered in deep scattering tissues such as the mouse brain, and permits AO correction over large axial fields of view. We demonstrate near-diffraction-limited imaging of deep cortical spines and (sub)cortical dendrites up to a depth of 1.4 mm (the edge of the mouse CA1 hippocampus). In addition, we show applications to deep-layer calcium imaging of astrocytes, including fibrous astrocytes that reside in the highly scattering corpus callosum.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

