The first wave of the COVID-19 epidemic in Spain was associated with early introductions and fast spread of a dominating genetic variant
- PMID: 34594042
- PMCID: PMC8481935
- DOI: 10.1038/s41588-021-00936-6
The first wave of the COVID-19 epidemic in Spain was associated with early introductions and fast spread of a dominating genetic variant
Abstract
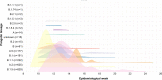
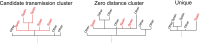
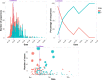
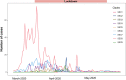
The coronavirus disease 2019 (COVID-19) pandemic has affected the world radically since 2020. Spain was one of the European countries with the highest incidence during the first wave. As a part of a consortium to monitor and study the evolution of the epidemic, we sequenced 2,170 samples, diagnosed mostly before lockdown measures. Here, we identified at least 500 introductions from multiple international sources and documented the early rise of two dominant Spanish epidemic clades (SECs), probably amplified by superspreading events. Both SECs were related closely to the initial Asian variants of SARS-CoV-2 and spread widely across Spain. We inferred a substantial reduction in the effective reproductive number of both SECs due to public-health interventions (Re < 1), also reflected in the replacement of SECs by a new variant over the summer of 2020. In summary, we reveal a notable difference in the initial genetic makeup of SARS-CoV-2 in Spain compared with other European countries and show evidence to support the effectiveness of lockdown measures in controlling virus spread, even for the most successful genetic variants.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Enfermedad por el coronavirus, situación en Españahttps://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual... (2020).
-
- Ministerio de la Presidencia, R. C. las C. y. M. D. BOE-A-2020-3692 https://www.boe.es/eli/es/rd/2020/03/14/463/con (2020).
Publication types
MeSH terms
Grants and funding
- 638553/ERC_/European Research Council/International
- SEJI/2019/011/Generalitat Valenciana (Regional Government of Valencia)
- StG 638553/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

