Sequencing the Complete Genome of COVID-19 Virus from Clinical Samples Using the Sanger Method
- PMID: 34594676
- PMCID: PMC8393061
- DOI: 10.46234/ccdcw2020.088
Sequencing the Complete Genome of COVID-19 Virus from Clinical Samples Using the Sanger Method
Abstract
What is already known on this topic?
Coronavirus disease 2019 (COVID-19), a disease caused by a novel human coronavirus named the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or COVID-19 virus, was reported in December 2019. Complete genomes of the COVID-19 virus from clinical samples using next generation sequencing (NGS) have been reported.
What is added by this report?
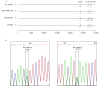
Here we provide the technical data for sequencing complete genome of COVID-19 virus from clinical samples using the Sanger method. Two complete COVID-19 virus genome sequences (named WH19004-S and GX0002) were obtained from clinical samples of COVID-19 patients, and two single nucleotide polymorphisms (SNPs) in ORF7a (T/C, nt 27,493) and ORF8 (T/C, nt 28,253) of WH19004-S were identified by Sanger sequencing.
What are the implications for public health practice?
The COVID-19 virus genome sequencing by Sanger method reported here could be used to generate data of high enough quality without requirement for expensive NGS equipment, which support sequencing complete genomes from clinical samples and monitoring of viral genetic variations of COVID-19 infections.
Copyright and License information: Editorial Office of CCDCW, Chinese Center for Disease Control and Prevention 2020.
Conflict of interest statement
Conflict of interest: No conflicts of interest were reported.
Figures

References
-
-
Tan WJ, Zhao X, Ma XJ, Wang WL, Niu PH, XU WB, et al. Notes from the field: a novel coronavirus genome identified in a cluster of pneumonia cases-Wuhan, China 2019−2020. China CDC Weekly 2020; 2(4): 61-2. http://weekly.chinacdc.cn/en/article/id/a3907201-f64f-4154-a19e-4253b453d10c.
-
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
