Assessing the accuracy of contact and distance predictions in CASP14
- PMID: 34595772
- PMCID: PMC8616839
- DOI: 10.1002/prot.26248
Assessing the accuracy of contact and distance predictions in CASP14
Abstract
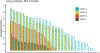
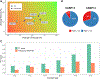
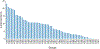
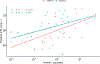
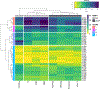
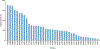
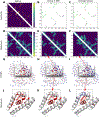
We present the results of the assessment of the intramolecular residue-residue contact and distance predictions from groups participating in the 14th round of the CASP experiment. The performance of contact prediction methods was evaluated with the measures used in previous CASPs, while distance predictions were assessed based on a new protocol, which considers individual distance pairs as well as the whole predicted distance matrix, using a graph-based framework. The results of the evaluation indicate that predictions by the tFold framework, TripletRes and DeepPotential were the most accurate in both categories. With regards to progress in method performance, the results of the assessment in contact prediction did not reveal any discernible difference when compared to CASP13. Arguably, this could be due to CASP14 FM targets being more challenging than ever before.
Keywords: CASP14; benchmarkin; community-wide experiment; numerical evaluation measures; prediction of residue-residue contact and distance.
© 2021 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Figures



References
-
- Göbel U, Sander C, Schneider R, et al. Correlated mutations and residue contacts in proteins. Proteins 1994;18:309–317. - PubMed
-
- de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet 2013;14:249–261. - PubMed
-
- Lesk AM. CASP2: report on ab initio predictions. Proteins 1997;Suppl 1:151–166. - PubMed
-
- Havel TF, Crippen GM, Kuntz ID. Effects of distance constraints on macromolecular conformation. II. Simulation of experimental results and theoretical predictions. Biopolymers 1979;18:73–81. 10.1002/bip.1979.360180108. - DOI

