Temporal spread and evolution of SARS-CoV-2 in the second pandemic wave in Brazil
- PMID: 34596904
- PMCID: PMC8661965
- DOI: 10.1002/jmv.27371
Temporal spread and evolution of SARS-CoV-2 in the second pandemic wave in Brazil
Abstract
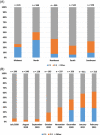
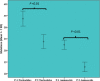
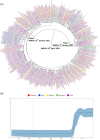
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic spread rapidly and this scenario is concerning in South America, mainly in Brazil that presented more than 21 million coronavirus disease 2019 cases and 590 000 deaths. The recent emergence of novel lineages carrying several mutations in the spike protein has raised additional public health concerns worldwide. The present study describes the temporal spreading and evolution of SARS-CoV2 in the beginning of the second pandemic wave in Brazil, highlighting the fast dissemination of the two major concerning variants (P.1 and P.2). A total of 2507 SARS-CoV-2 whole-genome sequences (WGSs) with available information from the country (Brazil) and sampling date (July 2020-February 2021), were obtained and the frequencies of the lineages were evaluated in the period of the growing second pandemic wave. The results demonstrated the increasing prevalence of P.1 and P.2 lineages in the period evaluated. P.2 lineage was first detected in the middle of 2020, but a high increase occurred only in the last trimester of this same year and the spreading to all Brazilian regions. P.1 lineage emerged even later, first in the North region in December 2020 and really fast dissemination to all other Brazilian regions in January and February 2021. All SARS-CoV-2 WGSs of P.1 and P.2 were further separately evaluated with a Bayesian approach. The rates of nucleotide and amino acid substitutions were statistically higher in P.1 than P.2 (p < 0.01). The phylodynamic analysis demonstrated that P.2 gradually spread in all the country from September 2020 to January 2021, while P.1 disseminated even faster from December 2020 to February 2021. Skyline plots of both lineages demonstrated a slight rise in the spreading for P.2 and exponential growth for P.1. In conclusion, these data demonstrated that the P.1 (recently renamed as Gamma) and P.2 lineages have predominated in the second pandemic wave due to the very high spreading across all geographic regions in Brazil at the end of 2020 and beginning of 2021.
Keywords: SARS coronavirus; dissemination; pandemic.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures


References
-
- Faria NR, Claro IM, Candido D, et al. Genomic characterisation of an emergent SARS‐CoV‐2 lineage in Manaus: preliminary findings. Virological. Accessed June 6, 2021. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-c...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

