Driving Single Cell Proteomics Forward with Innovation
- PMID: 34597050
- PMCID: PMC8571067
- DOI: 10.1021/acs.jproteome.1c00639
Driving Single Cell Proteomics Forward with Innovation
Abstract
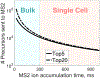
Current single-cell mass spectrometry (MS) methods can quantify thousands of peptides per single cell while detecting peptide-like features that may support the quantification of 10-fold more peptides. This 10-fold gain might be attained by innovations in data acquisition and interpretation even while using existing instrumentation. This perspective discusses possible directions for such innovations with the aim to stimulate community efforts for increasing the coverage and quantitative accuracy of single proteomics while simultaneously decreasing missing data. Parallel improvements in instrumentation, sample preparation, and peptide separation will afford additional gains. Together, these synergistic routes for innovation project a rapid growth in the capabilities of MS based single-cell protein analysis. These gains will directly empower applications of single-cell proteomics to biomedical research.
Keywords: data acquisition; data interpretation; peptide identity propagation; single-cell proteomics; ultrasensitive proteomics.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

