A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response
- PMID: 34597124
- PMCID: PMC8593910
- DOI: 10.1126/sciimmunol.abg7836
A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response
Abstract
“Stem-like” TCF1+ CD8+ T (TSL) cells are necessary for long-term maintenance of T cell responses and the efficacy of immunotherapy, but, as tumors contain signals that should drive T cell terminal differentiation, how these cells are maintained in tumors remains unclear. In this study, we found that a small number of TCF1+ tumor-specific CD8+ T cells were present in lung tumors throughout their development. Yet, most intratumoral T cells differentiated as tumors progressed, corresponding with an immunologic shift in the tumor microenvironment (TME) from “hot” (T cell inflamed) to “cold” (non–T cell inflamed). By contrast, most tumor-specific CD8+ T cells in tumor-draining lymph nodes (dLNs) had functions and gene expression signatures similar to TSL from chronic lymphocytic choriomeningitis virus infection, and this population was stable over time despite the changes in the TME. dLN T cells were the developmental precursors of, and were clonally related to, their more differentiated intratumoral counterparts. Our data support the hypothesis that dLN T cells are the developmental precursors of the TCF1+ T cells in tumors that are maintained by continuous migration. Last, CD8+ T cells similar to TSL were also present in LNs from patients with lung adenocarcinoma, suggesting that a similar model may be relevant in human disease. Thus, we propose that the dLN TSL reservoir has a critical function in sustaining antitumor T cells during tumor development and in protecting them from the terminal differentiation that occurs in the TME.
Conflict of interest statement
Figures

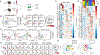
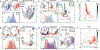
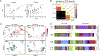
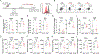

References
-
- Duma N, Santana-Davila R, Molina JR, Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 94, 1623–1640 (2019). - PubMed
-
- Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HEK, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS, Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). - PMC - PubMed
-
- Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L, Colocalization of Inflammatory Response with B7-H1 Expression in Human Melanocytic Lesions Supports an Adaptive Resistance Mechanism of Immune Escape. Sci Transl Med 4, (2012). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- T32 CA233414/CA/NCI NIH HHS/United States
- K22 CA200912/CA/NCI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- T32 GM080202/GM/NIGMS NIH HHS/United States
- P30 CA016359/CA/NCI NIH HHS/United States
- T32 HL007974/HL/NHLBI NIH HHS/United States
- P50 CA196530/CA/NCI NIH HHS/United States
- R01 AI125741/AI/NIAID NIH HHS/United States
- T32 AI007019/AI/NIAID NIH HHS/United States
- T32 AR007016/AR/NIAMS NIH HHS/United States
- F30 DK127526/DK/NIDDK NIH HHS/United States
- R01 AI148403/AI/NIAID NIH HHS/United States
- T32 AI155387/AI/NIAID NIH HHS/United States
- R01 CA237037/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

