GABARAP sequesters the FLCN-FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis
- PMID: 34597140
- PMCID: PMC10938568
- DOI: 10.1126/sciadv.abj2485
GABARAP sequesters the FLCN-FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis
Abstract
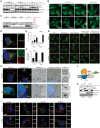
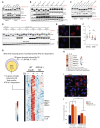
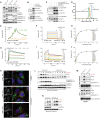
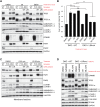
Adaptive changes in lysosomal capacity are driven by the transcription factors TFEB and TFE3 in response to increased autophagic flux and endolysosomal stress, yet the molecular details of their activation are unclear. LC3 and GABARAP members of the ATG8 protein family are required for selective autophagy and sensing perturbation within the endolysosomal system. Here, we show that during the conjugation of ATG8 to single membranes (CASM), Parkin-dependent mitophagy, and Salmonella-induced xenophagy, the membrane conjugation of GABARAP, but not LC3, is required for activation of TFEB/TFE3 to control lysosomal capacity. GABARAP directly binds to a previously unidentified LC3-interacting motif (LIR) in the FLCN/FNIP tumor suppressor complex and mediates sequestration to GABARAP-conjugated membrane compartments. This disrupts FLCN/FNIP GAP function toward RagC/D, resulting in impaired substrate-specific mTOR-dependent phosphorylation of TFEB. Thus, the GABARAP-FLCN/FNIP-TFEB axis serves as a molecular sensor that coordinates lysosomal homeostasis with perturbations and cargo flux within the autophagy-lysosomal network.
Figures


References
-
- Dikic I., Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017). - PubMed
-
- Lawrence R. E., Zoncu R., The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019). - PubMed
-
- Kaur J., Debnath J., Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472 (2015). - PubMed
-
- Ballabio A., Bonifacino J. S., Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2020). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

