Radiomics in Oncology: A Practical Guide
- PMID: 34597235
- PMCID: PMC8501897
- DOI: 10.1148/rg.2021210037
Radiomics in Oncology: A Practical Guide
Abstract
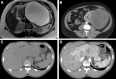
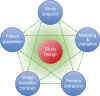
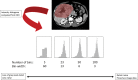
Radiomics refers to the extraction of mineable data from medical imaging and has been applied within oncology to improve diagnosis, prognostication, and clinical decision support, with the goal of delivering precision medicine. The authors provide a practical approach for successfully implementing a radiomic workflow from planning and conceptualization through manuscript writing. Applications in oncology typically are either classification tasks that involve computing the probability of a sample belonging to a category, such as benign versus malignant, or prediction of clinical events with a time-to-event analysis, such as overall survival. The radiomic workflow is multidisciplinary, involving radiologists and data and imaging scientists, and follows a stepwise process involving tumor segmentation, image preprocessing, feature extraction, model development, and validation. Images are curated and processed before segmentation, which can be performed on tumors, tumor subregions, or peritumoral zones. Extracted features typically describe the distribution of signal intensities and spatial relationship of pixels within a region of interest. To improve model performance and reduce overfitting, redundant and nonreproducible features are removed. Validation is essential to estimate model performance in new data and can be performed iteratively on samples of the dataset (cross-validation) or on a separate hold-out dataset by using internal or external data. A variety of noncommercial and commercial radiomic software applications can be used. Guidelines and artificial intelligence checklists are useful when planning and writing up radiomic studies. Although interest in the field continues to grow, radiologists should be familiar with potential pitfalls to ensure that meaningful conclusions can be drawn. Online supplemental material is available for this article. Published under a CC BY 4.0 license.
Conflict of interest statement
Figures




References
-
- Lambin P , Leijenaar RTH , Deist TM , et al. . Radiomics: the bridge between medical imaging and personalized medicine . Nat Rev Clin Oncol 2017. ; 14 ( 12 ): 749 – 762 . - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

