Domestication Shapes Recombination Patterns in Tomato
- PMID: 34597400
- PMCID: PMC8763028
- DOI: 10.1093/molbev/msab287
Domestication Shapes Recombination Patterns in Tomato
Abstract
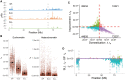
Meiotic recombination is a biological process of key importance in breeding, to generate genetic diversity and develop novel or agronomically relevant haplotypes. In crop tomato, recombination is curtailed as manifested by linkage disequilibrium decay over a longer distance and reduced diversity compared with wild relatives. Here, we compared domesticated and wild populations of tomato and found an overall conserved recombination landscape, with local changes in effective recombination rate in specific genomic regions. We also studied the dynamics of recombination hotspots resulting from domestication and found that loss of such hotspots is associated with selective sweeps, most notably in the pericentromeric heterochromatin. We detected footprints of genetic changes and structural variants, among them associated with transposable elements, linked with hotspot divergence during domestication, likely causing fine-scale alterations to recombination patterns and resulting in linkage drag.
Keywords: domestication; heterochromatin; recombination; selective sweeps; structural variants; transposable elements.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Aflitos S, Schijlen E, de Jong H, de Ridder D, Smit S, Finkers R, Wang J, Zhang G, Li N, Mao L, 100 Tomato Genome Sequencing Consortium, et al.2014. Exploring genetic variation in the tomato (Solanum section lycopersicon) clade by whole-genome sequencing. Plant J. 80(1):136–148. - PubMed
-
- Allard RW. 1999. History of plant population genetics. Annu Rev Genet. 33:1–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

