eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale
- PMID: 34597405
- PMCID: PMC8662613
- DOI: 10.1093/molbev/msab293
eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale
Abstract
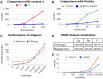
Even though automated functional annotation of genes represents a fundamental step in most genomic and metagenomic workflows, it remains challenging at large scales. Here, we describe a major upgrade to eggNOG-mapper, a tool for functional annotation based on precomputed orthology assignments, now optimized for vast (meta)genomic data sets. Improvements in version 2 include a full update of both the genomes and functional databases to those from eggNOG v5, as well as several efficiency enhancements and new features. Most notably, eggNOG-mapper v2 now allows for: 1) de novo gene prediction from raw contigs, 2) built-in pairwise orthology prediction, 3) fast protein domain discovery, and 4) automated GFF decoration. eggNOG-mapper v2 is available as a standalone tool or as an online service at http://eggnog-mapper.embl.de.
Keywords: bioinformatics; computational genomics; functional annotation; metagenomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

