Classification of 101 BRCA1 and BRCA2 variants of uncertain significance by cosegregation study: A powerful approach
- PMID: 34597585
- PMCID: PMC8546044
- DOI: 10.1016/j.ajhg.2021.09.003
Classification of 101 BRCA1 and BRCA2 variants of uncertain significance by cosegregation study: A powerful approach
Abstract
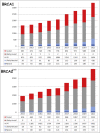
Up to 80% of BRCA1 and BRCA2 genetic variants remain of uncertain clinical significance (VUSs). Only variants classified as pathogenic or likely pathogenic can guide breast and ovarian cancer prevention measures and treatment by PARP inhibitors. We report the first results of the ongoing French national COVAR (cosegregation variant) study, the aim of which is to classify BRCA1/2 VUSs. The classification method was a multifactorial model combining different associations between VUSs and cancer, including cosegregation data. At this time, among the 653 variants selected, 101 (15%) distinct variants shared by 1,624 families were classified as pathogenic/likely pathogenic or benign/likely benign by the COVAR study. Sixty-six of the 101 (65%) variants classified by COVAR would have remained VUSs without cosegregation data. Of note, among the 34 variants classified as pathogenic by COVAR, 16 remained VUSs or likely pathogenic when following the ACMG/AMP variant classification guidelines. Although the initiation and organization of cosegregation analyses require a considerable effort, the growing number of available genetic tests results in an increasing number of families sharing a particular variant, and thereby increases the power of such analyses. Here we demonstrate that variant cosegregation analyses are a powerful tool for the classification of variants in the BRCA1/2 breast-ovarian cancer predisposition genes.
Keywords: BRCA1; BRCA2; classification; clinical; cosegregation data; multifactorial model; variant of uncertain significance.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.S.-L. and the Institut Curie have received honoraria for her participation in education meetings organized by AstraZeneca or Tesaro. The remaining authors declare no conflict of interest.
Figures


References
-
- Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., BRCA1 and BRCA2 Cohort Consortium Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. - PubMed
-
- Lesueur F., Mebirouk N., Jiao Y., Barjhoux L., Belotti M., Laurent M., Léone M., Houdayer C., Bressac-de Paillerets B., Vaur D., GEMO Study Collaborators GEMO, a National Resource to Study Genetic Modifiers of Breast and Ovarian Cancer Risk in BRCA1 and BRCA2 Pathogenic Variant Carriers. Front. Oncol. 2018;8:490. - PMC - PubMed
-
- Rouleau E., Jesson B., Briaux A., Nogues C., Chabaud V., Demange L., Sokolowska J., Coulet F., Barouk-Simonet E., Bignon Y.J. Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res. Treat. 2012;133:1179–1190. - PubMed
-
- Caputo S., Benboudjema L., Sinilnikova O., Rouleau E., Béroud C., Lidereau R., French BRCA GGC Consortium Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40:D992–D1002. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

