The Protexin complex counters resection on stalled forks to promote homologous recombination and crosslink repair
- PMID: 34597596
- PMCID: PMC8588999
- DOI: 10.1016/j.molcel.2021.09.008
The Protexin complex counters resection on stalled forks to promote homologous recombination and crosslink repair
Abstract
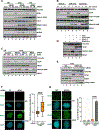
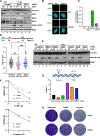
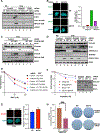
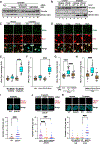
Protection of stalled replication forks is critical to genomic stability. Using genetic and proteomic analyses, we discovered the Protexin complex containing the ssDNA binding protein SCAI and the DNA polymerase REV3. Protexin is required specifically for protecting forks stalled by nucleotide depletion, fork barriers, fragile sites, and DNA inter-strand crosslinks (ICLs), where it promotes homologous recombination and repair. Protexin loss leads to ssDNA accumulation and profound genomic instability in response to ICLs. Protexin interacts with RNA POL2, and both oppose EXO1's resection of DNA on forks remodeled by the FANCM translocase activity. This pathway acts independently of BRCA/RAD51-mediated fork stabilization, and cells with BRCA2 mutations were dependent on SCAI for survival. These data suggest that Protexin and its associated factors establish a new fork protection pathway that counteracts fork resection in part through a REV3 polymerase-dependent resynthesis mechanism of excised DNA, particularly at ICL stalled forks.
Keywords: CRISPR; EXO1; FANCM; Protexin; REV3L; SCAI; homologous recombination; inter-strand crosslinks; replication stress; resection.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests. S.J.E. is a member of the Molecular Cell advisory board.
Figures



References
-
- Brandt DT, Baarlink C, Kitzing TM, Kremmer E, Ivaska J, Nollau P, and Grosse R (2009). SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol 11, 557–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

