Tagmentation-based single-cell genomics
- PMID: 34599003
- PMCID: PMC8494221
- DOI: 10.1101/gr.275223.121
Tagmentation-based single-cell genomics
Abstract
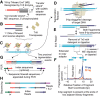
It has been just over 10 years since the initial description of transposase-based methods to prepare high-throughput sequencing libraries, or "tagmentation," in which a hyperactive transposase is used to simultaneously fragment target DNA and append universal adapter sequences. Tagmentation effectively replaced a series of processing steps in traditional workflows with one single reaction. It is the simplicity, coupled with the high efficiency of tagmentation, that has made it a favored means of sequencing library construction and fueled a diverse range of adaptations to assay a variety of molecular properties. In recent years, this has been centered in the single-cell space with a catalog of tagmentation-based assays that have been developed, covering a substantial swath of the regulatory landscape. To date, there have been a number of excellent reviews on single-cell technologies structured around the molecular properties that can be profiled. This review is instead framed around the central components and properties of tagmentation and how they have enabled the development of innovative molecular tools to probe the regulatory landscape of single cells. Furthermore, the granular specifics on cell throughput or richness of data provided by the extensive list of individual technologies are not discussed. Such metrics are rapidly changing and highly sample specific and are better left to studies that directly compare technologies for assays against one another in a rigorously controlled framework. The hope for this review is that, in laying out the diversity of molecular techniques at each stage of these assay platforms, new ideas may arise for others to pursue that will further advance the field of single-cell genomics.
© 2021 Adey; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- 10x Genomics. 2021. Multiome ATAC + gene expression. Pleasanton, CA. https://www.10xgenomics.com/products/single-cell-multiome-atac-plus-gene....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
