Nanoconfinement of microvilli alters gene expression and boosts T cell activation
- PMID: 34599101
- PMCID: PMC8501847
- DOI: 10.1073/pnas.2107535118
Nanoconfinement of microvilli alters gene expression and boosts T cell activation
Erratum in
-
Correction for Aramesh et al., Nanoconfinement of microvilli alters gene expression and boosts T cell activation.Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2202699119. doi: 10.1073/pnas.2202699119. Epub 2022 Apr 1. Proc Natl Acad Sci U S A. 2022. PMID: 35363570 Free PMC article. No abstract available.
Abstract
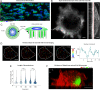
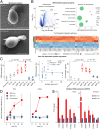
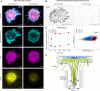
T cells sense and respond to their local environment at the nanoscale by forming small actin-rich protrusions, called microvilli, which play critical roles in signaling and antigen recognition, particularly at the interface with the antigen presenting cells. However, the mechanism by which microvilli contribute to cell signaling and activation is largely unknown. Here, we present a tunable engineered system that promotes microvilli formation and T cell signaling via physical stimuli. We discovered that nanoporous surfaces favored microvilli formation and markedly altered gene expression in T cells and promoted their activation. Mechanistically, confinement of microvilli inside of nanopores leads to size-dependent sorting of membrane-anchored proteins, specifically segregating CD45 phosphatases and T cell receptors (TCR) from the tip of the protrusions when microvilli are confined in 200-nm pores but not in 400-nm pores. Consequently, formation of TCR nanoclustered hotspots within 200-nm pores allows sustained and augmented signaling that prompts T cell activation even in the absence of TCR agonists. The synergistic combination of mechanical and biochemical signals on porous surfaces presents a straightforward strategy to investigate the role of microvilli in T cell signaling as well as to boost T cell activation and expansion for application in the growing field of adoptive immunotherapy.
Keywords: T cell microvilli; cell-surface interactions; immunoengineering; mechanobiology; nanoconfinement.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Aramesh M., et al., Functionalized bead assay to measure three-dimensional traction forces during T-cell activation. Nano Lett. 21, 507–514 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

