Listeriolysin S: A bacteriocin from Listeria monocytogenes that induces membrane permeabilization in a contact-dependent manner
- PMID: 34599102
- PMCID: PMC8501752
- DOI: 10.1073/pnas.2108155118
Listeriolysin S: A bacteriocin from Listeria monocytogenes that induces membrane permeabilization in a contact-dependent manner
Abstract
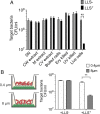
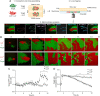
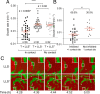
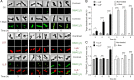
Listeriolysin S (LLS) is a thiazole/oxazole-modified microcin (TOMM) produced by hypervirulent clones of Listeria monocytogenes LLS targets specific gram-positive bacteria and modulates the host intestinal microbiota composition. To characterize the mechanism of LLS transfer to target bacteria and its bactericidal function, we first investigated its subcellular distribution in LLS-producer bacteria. Using subcellular fractionation assays, transmission electron microscopy, and single-molecule superresolution microscopy, we identified that LLS remains associated with the bacterial cell membrane and cytoplasm and is not secreted to the bacterial extracellular space. Only living LLS-producer bacteria (and not purified LLS-positive bacterial membranes) display bactericidal activity. Applying transwell coculture systems and microfluidic-coupled microscopy, we determined that LLS requires direct contact between LLS-producer and -target bacteria in order to display bactericidal activity, and thus behaves as a contact-dependent bacteriocin. Contact-dependent exposure to LLS leads to permeabilization/depolarization of the target bacterial cell membrane and adenosine triphosphate (ATP) release. Additionally, we show that lipoteichoic acids (LTAs) can interact with LLS and that LTA decorations influence bacterial susceptibility to LLS. Overall, our results suggest that LLS is a TOMM that displays a contact-dependent inhibition mechanism.
Keywords: Listeria monocytogenes (Lm); Listeriolysin S (LLS); bacteriocin; contact-dependent inhibition (CDI); microfluidic microscopy.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Radoshevich L., Cossart P., Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46 (2018). - PubMed
-
- Orsi R. H., den Bakker H. C., Wiedmann M., Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

