Applying and improving AlphaFold at CASP14
- PMID: 34599769
- PMCID: PMC9299164
- DOI: 10.1002/prot.26257
Applying and improving AlphaFold at CASP14
Abstract
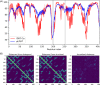
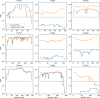
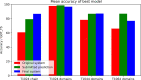
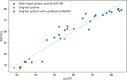
We describe the operation and improvement of AlphaFold, the system that was entered by the team AlphaFold2 to the "human" category in the 14th Critical Assessment of Protein Structure Prediction (CASP14). The AlphaFold system entered in CASP14 is entirely different to the one entered in CASP13. It used a novel end-to-end deep neural network trained to produce protein structures from amino acid sequence, multiple sequence alignments, and homologous proteins. In the assessors' ranking by summed z scores (>2.0), AlphaFold scored 244.0 compared to 90.8 by the next best group. The predictions made by AlphaFold had a median domain GDT_TS of 92.4; this is the first time that this level of average accuracy has been achieved during CASP, especially on the more difficult Free Modeling targets, and represents a significant improvement in the state of the art in protein structure prediction. We reported how AlphaFold was run as a human team during CASP14 and improved such that it now achieves an equivalent level of performance without intervention, opening the door to highly accurate large-scale structure prediction.
Keywords: AlphaFold; CASP; deep learning; machine learning; protein structure prediction.
© 2021 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
John Jumper, Richard Evans, Alexander Pritzel, Tim Green, Michael Figurnov, Olaf Ronneberger, Russ Bates, Alex Bridgland, Simon A. A. Kohl, David Reiman, and Andrew W. Senior have filed provisional patent applications relating to machine learning for predicting protein structures. The remaining authors declare no competing interests.
Figures

References
-
- Senior AW, Evans R, Jumper J, et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706‐710. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

