Genome-wide phenotypic RNAi screen in the Drosophila wing: global parameters
- PMID: 34599819
- PMCID: PMC8962446
- DOI: 10.1093/g3journal/jkab351
Genome-wide phenotypic RNAi screen in the Drosophila wing: global parameters
Abstract
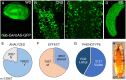
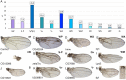
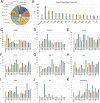
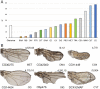
We have screened a collection of UAS-RNAi lines targeting 10,920 Drosophila protein-coding genes for phenotypes in the adult wing. We identified 3653 genes (33%) whose knockdown causes either larval/pupal lethality or a mutant phenotype affecting the formation of a normal wing. The most frequent phenotypes consist of changes in wing size, vein differentiation, and patterning, defects in the wing margin and in the apposition of the dorsal and ventral wing surfaces. We also defined 16 functional categories encompassing the most relevant aspect of each protein function and assigned each Drosophila gene to one of these functional groups. This allowed us to identify which mutant phenotypes are enriched within each functional group. Finally, we used previously published gene expression datasets to determine which genes are or are not expressed in the wing disc. Integrating expression, phenotypic and molecular information offers considerable precision to identify the relevant genes affecting wing formation and the biological processes regulated by them.
Keywords: RNAi; phenotype; screen; wing.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures



References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al.2000. The genome sequence of Drosophila melanogaster. Science. 287:2185–2195. - PubMed
-
- Bao R, Dia SE, Issa HA, Alhusein D, Friedrich M.. 2018. Comparative evidence of an exceptional impact of gene duplication on the developmental evolution of Drosophila and the higher diptera. Front Ecol Evol. 6:63.doi:10.3389/fevo.2018.00063.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
