Cytokine storm in the pathophysiology of COVID-19: Possible functional disturbances of miRNAs
- PMID: 34601331
- PMCID: PMC8452524
- DOI: 10.1016/j.intimp.2021.108172
Cytokine storm in the pathophysiology of COVID-19: Possible functional disturbances of miRNAs
Abstract
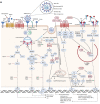
SARS-CoV-2, as the causative agent of COVID-19, is an enveloped positives-sense single-stranded RNA virus that belongs to the Beta-CoVs sub-family. A sophisticated hyper-inflammatory reaction named cytokine storm is occurred in patients with severe/critical COVID-19, following an imbalance in immune-inflammatory processes and inhibition of antiviral responses by SARS-CoV-2, which leads to pulmonary failure, ARDS, and death. The miRNAs are small non-coding RNAs with an average length of 22 nucleotides which play various roles as one of the main modulators of genes expression and maintenance of immune system homeostasis. Recent evidence has shown that Homo sapiens (hsa)-miRNAs have the potential to work in three pivotal areas including targeting the virus genome, regulating the inflammatory signaling pathways, and reinforcing the production/signaling of IFNs-I. However, it seems that several SARS-CoV-2-induced interfering agents such as viral (v)-miRNAs, cytokine content, competing endogenous RNAs (ceRNAs), etc. preclude efficient function of hsa-miRNAs in severe/critical COVID-19. This subsequently leads to increased virus replication, intense inflammatory processes, and secondary complications development. In this review article, we provide an overview of hsa-miRNAs roles in viral genome targeting, inflammatory pathways modulation, and IFNs responses amplification in severe/critical COVID-19 accompanied by probable interventional factors and their function. Identification and monitoring of these interventional elements can help us in designing the miRNAs-based therapy for the reduction of complications/mortality rate in patients with severe/critical forms of the disease.
Keywords: COVID-19; Cytokine storm; Gene regulators; SARS-CoV-2; miRNAs; miRNAs-based therapy.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

