Antimicrobial Resistance and CRISPR Typing Among Salmonella Isolates From Poultry Farms in China
- PMID: 34603259
- PMCID: PMC8481896
- DOI: 10.3389/fmicb.2021.730046
Antimicrobial Resistance and CRISPR Typing Among Salmonella Isolates From Poultry Farms in China
Abstract
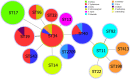
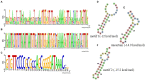
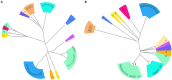
Although knowledge of the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas system has been applied in many research areas, comprehensive studies of this system in Salmonella, particularly in analysis of antibiotic resistance, have not been reported. In this work, 75 Salmonella isolates obtained from broilers or broilers products were characterized to determine their antimicrobial susceptibilities, antibiotic resistance gene profiles, and CRISPR array diversities, and genotyping was explored. In total, 80.00% (60/75) of the strains were multidrug resistant, and the main pattern observed in the isolates was CN-AZM-AMP-AMC-CAZ-CIP-ATM-TE-SXT-FOS-C. The resistance genes of streptomycin (aadA), phenicol (floR-like and catB3-like), β-lactams (bla TEM, bla OXA, and bla CTX), tetracycline [tet(A)-like], and sulfonamides (sul1 and sul2) appeared at higher frequencies among the corresponding resistant isolates. Subsequently, we analyzed the CRISPR arrays and found 517 unique spacer sequences and 31 unique direct repeat sequences. Based on the CRISPR spacer sequences, we developed a novel typing method, CRISPR locus three spacer sequences typing (CLTSST), to help identify sources of Salmonella outbreaks especially correlated with epidemiological data. Compared with multi-locus sequence typing (MLST), conventional CRISPR typing (CCT), and CRISPR locus spacer pair typing (CLSPT), discrimination using CLTSST was weaker than that using CCT but stronger than that using MLST and CLSPT. In addition, we also found that there were no close correlations between CRISPR loci and antibiotics but had close correlations between CRISPR loci and antibiotic resistance genes in Salmonella isolates.
Keywords: CRISPR array; CRISPR-Cas; Salmonella; antimicrobial resistance; molecular typing.
Copyright © 2021 Li, Wang, Gao, Li, Ma and Wang.
Conflict of interest statement
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

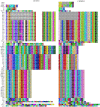
Similar articles
-
New clustered regularly interspaced short palindromic repeat locus spacer pair typing method based on the newly incorporated spacer for Salmonella enterica.J Clin Microbiol. 2014 Aug;52(8):2955-62. doi: 10.1128/JCM.00696-14. Epub 2014 Jun 4. J Clin Microbiol. 2014. PMID: 24899040 Free PMC article.
-
Genetic analysis and CRISPR typing of Salmonella enterica serovar Enteritidis from different sources revealed potential transmission from poultry and pig to human.Int J Food Microbiol. 2018 Feb 2;266:119-125. doi: 10.1016/j.ijfoodmicro.2017.11.025. Epub 2017 Nov 28. Int J Food Microbiol. 2018. PMID: 29212058
-
Comprehensive Assessment of Subtyping Methods for Improved Surveillance of Foodborne Salmonella.Microbiol Spectr. 2022 Oct 26;10(5):e0247922. doi: 10.1128/spectrum.02479-22. Epub 2022 Oct 4. Microbiol Spectr. 2022. PMID: 36194132 Free PMC article.
-
Prevalence and antimicrobial resistance of Salmonellaisolates from goose farms in Northeast China.Iran J Vet Res. 2020 Fall;21(4):287-293. Iran J Vet Res. 2020. PMID: 33584841 Free PMC article.
-
Clustered Regularly Interspaced Short Palindromic Repeats Genotyping of Multidrug-Resistant Salmonella Heidelberg Strains Isolated From the Poultry Production Chain Across Brazil.Front Microbiol. 2022 Jun 17;13:867278. doi: 10.3389/fmicb.2022.867278. eCollection 2022. Front Microbiol. 2022. PMID: 35783410 Free PMC article.
Cited by
-
Large-Scale Phylogenetic Analysis Reveals a New Genetic Clade among Escherichia coli O26 Strains.Microbiol Spectr. 2022 Feb 23;10(1):e0252521. doi: 10.1128/spectrum.02525-21. Epub 2022 Feb 2. Microbiol Spectr. 2022. PMID: 35107330 Free PMC article.
-
One Health: a holistic approach for food safety in livestock.Sci One Health. 2023 May 10;1:100015. doi: 10.1016/j.soh.2023.100015. eCollection 2022 Nov. Sci One Health. 2023. PMID: 39076604 Free PMC article. Review.
-
Phenotypic and genotypic characterization of antimicrobial resistance profiles in Salmonella isolated from waterfowl in 2002-2005 and 2018-2020 in Sichuan, China.Front Microbiol. 2022 Oct 6;13:987613. doi: 10.3389/fmicb.2022.987613. eCollection 2022. Front Microbiol. 2022. PMID: 36274743 Free PMC article.
-
Prevalence and Characterization of Salmonella Isolated from Chickens in Anhui, China.Pathogens. 2023 Mar 16;12(3):465. doi: 10.3390/pathogens12030465. Pathogens. 2023. PMID: 36986387 Free PMC article.
-
Sub-MIC antibiotics increased the fitness cost of CRISPR-Cas in Acinetobacter baumannii.Front Microbiol. 2024 Jul 1;15:1381749. doi: 10.3389/fmicb.2024.1381749. eCollection 2024. Front Microbiol. 2024. PMID: 39011146 Free PMC article.
References
-
- Best E. L., Hampton M. D., Ethelberg S., Liebana E., Clifton-Hadley F. A., Threlfall E. J. (2009). Drug-resistant salmonella Typhimurium DT 120: use of PFGE and MLVA in a putative international outbreak investigation. Microb. Drug Resist. 15, 133–138. doi: 10.1089/mdr.2009.0911, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

