A Systematic Narration of Some Key Concepts and Procedures in Plant Breeding
- PMID: 34603352
- PMCID: PMC8481876
- DOI: 10.3389/fpls.2021.724517
A Systematic Narration of Some Key Concepts and Procedures in Plant Breeding
Abstract
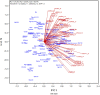
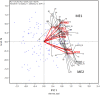
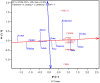
The goal of a plant breeding program is to develop new cultivars of a crop kind with improved yield and quality for a target region and end-use. Improved yield across locations and years means better adaptation to the climatic, soil, and management conditions in the target region. Improved or maintained quality renders and adds value to the improved yield. Both yield and quality must be considered simultaneously, which constitutes the greatest challenge to successful cultivar development. Cultivar development consists of two stages: the development of a promising breeding population and the selection of the best genotypes out of it. A complete breeder's equation was presented to cover both stages, which consists of three key parameters for a trait of interest: the population mean (μ), the population variability (σ G ), and the achieved heritability (h 2 or H), under the multi-location, multi-year framework. Population development is to maximize μσ G and progeny selection is to improve H. Approaches to improve H include identifying and utilizing repeatable genotype by environment interaction (GE) through mega-environment analysis, accommodating unrepeatable GE through adequate testing, and reducing experimental error via replication and spatial analysis. Related concepts and procedures were critically reviewed, including GGE (genotypic main effect plus genotype by environment interaction) biplot analysis, GGE + GGL (genotypic main effect plus genotype by location interaction) biplot analysis, LG (location-grouping) biplot analysis, stability analysis, spatial analysis, adequate testing, and optimum replication. Selection on multiple traits includes independent culling and index selection, for the latter GYT (genotype by yield*trait) biplot analysis was recommended. Genomic selection may provide an alternative and potentially more effective approach in all these aspects. Efforts were made to organize and comment on these concepts and procedures in a systematic manner.
Keywords: biplot analysis; breeder's equation; genotype by environment interaction; heritability; mega-environment analysis; multi-trait selection; optimum replication; optimum testing.
Copyright © 2021 Yan.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Alwala S., Kwolek T., McPherson M., Pellow J., Meyer D. (2010). A comprehensive comparison between Eberhart and Russell joint regression and GGE biplot analyses to identify stable and high yielding maize hybrids. Field Crops Res. 119, 225–230. 10.1016/j.fcr.2010.07.010 - DOI
-
- Amira J. O., Ojo D. K., Ariyo O. J., Oduwaye O. A., Ayo-Vaughan M. A. (2013). Relative discriminating powers of GGE and AMMI models in the selection of tropical soybean genotypes. Afr. Crop Sci. J. 21, 67–73.
-
- Annicchiarico P. (2021). Breeding gain from exploitation of regional adaptation: an Alfalfa case study. Crop Sci. 61, 2254–2271. 10.1002/csc2.20423 - DOI
-
- Arief V. N., DeLacy I. H., Crossa J., Payne T., Singh R., Braun H. J., et al. (2015). Evaluating testing strategies for plant breeding field trials: redesigning a CIMMYT international wheat nursery. Crop Sci. 55, 164–177. 10.2135/cropsci2014.06.0415 - DOI
-
- Asins M. J. (2002). Present and future of quantitative trait locus analysis in plant breeding. Plant Breed. 121, 281–291. 10.1046/j.1439-0523.2002.730285.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

