Graph Representation Forecasting of Patient's Medical Conditions: Toward a Digital Twin
- PMID: 34603366
- PMCID: PMC8481902
- DOI: 10.3389/fgene.2021.652907
Graph Representation Forecasting of Patient's Medical Conditions: Toward a Digital Twin
Abstract
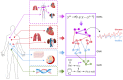
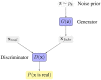
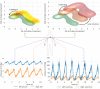
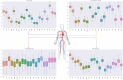
Objective: Modern medicine needs to shift from a wait and react, curative discipline to a preventative, interdisciplinary science aiming at providing personalized, systemic, and precise treatment plans to patients. To this purpose, we propose a "digital twin" of patients modeling the human body as a whole and providing a panoramic view over individuals' conditions. Methods: We propose a general framework that composes advanced artificial intelligence (AI) approaches and integrates mathematical modeling in order to provide a panoramic view over current and future pathophysiological conditions. Our modular architecture is based on a graph neural network (GNN) forecasting clinically relevant endpoints (such as blood pressure) and a generative adversarial network (GAN) providing a proof of concept of transcriptomic integrability. Results: We tested our digital twin model on two simulated clinical case studies combining information at organ, tissue, and cellular level. We provided a panoramic overview over current and future patient's conditions by monitoring and forecasting clinically relevant endpoints representing the evolution of patient's vital parameters using the GNN model. We showed how to use the GAN to generate multi-tissue expression data for blood and lung to find associations between cytokines conditioned on the expression of genes in the renin-angiotensin pathway. Our approach was to detect inflammatory cytokines, which are known to have effects on blood pressure and have previously been associated with SARS-CoV-2 infection (e.g., CXCR6, XCL1, and others). Significance: The graph representation of a computational patient has potential to solve important technological challenges in integrating multiscale computational modeling with AI. We believe that this work represents a step forward toward next-generation devices for precision and predictive medicine.
Keywords: digital twin; generative adversarial networks; graph representation learning; monitoring; precision medicine.
Copyright © 2021 Barbiero, Viñas Torné and Lió.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



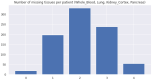
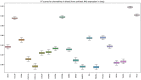

References
-
- Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., et al. (2016). Tensorflow: a system for large-scale machine learning, in 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16) (Savannah, GA: ), 265–283.
-
- Aguet F., Barbeira A. N., Bonazzola R., Brown A., Castel S. E., Jo B., et al. (2019). The gtex consortium atlas of genetic regulatory effects across human tissues. bioRxiv [Preprint]. 10.1101/787903 - DOI
-
- Arjovsky M., Chintala S., Bottou L. (2017). Wasserstein GAN. arXiv [Preprint]. arXiv:1701.07875.
-
- Barbiero P., Lió P. (2020). The computational patient has diabetes and a covid. arXiv [Preprint]. arXiv:2006.06435.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

