Integrative Analyses of Genes Associated With Right Ventricular Cardiomyopathy Induced by Tricuspid Regurgitation
- PMID: 34603374
- PMCID: PMC8485137
- DOI: 10.3389/fgene.2021.708275
Integrative Analyses of Genes Associated With Right Ventricular Cardiomyopathy Induced by Tricuspid Regurgitation
Abstract
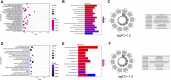
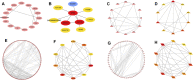
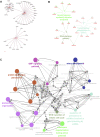
Tricuspid regurgitation (TR) induces right ventricular cardiomyopathy, a common heart disease, and eventually leads to severe heart failure and serious clinical complications. Accumulating evidence shows that long non-coding RNAs (lncRNAs) are involved in the pathological process of a variety of cardiovascular diseases. However, the regulatory mechanisms and functional roles of RNA interactions in TR-induced right ventricular cardiomyopathy are still unclear. Accordingly, we performed integrative analyses of genes associated with right ventricular cardiomyopathy induced by TR to study the roles of lncRNAs in the pathogenesis of this disease. In this study, we used high-throughput sequencing data of tissue samples from nine clinical cases of right ventricular myocardial cardiomyopathy induced by TR and nine controls with normal right ventricular myocardium from the Genotype-Tissue Expression database. We identified differentially expressed lncRNAs and constructed a protein-protein interaction and lncRNA-messenger RNA (mRNA) co-expression network. Furthermore, we determined hub lncRNA-mRNA modules related to right ventricular myocardial disease induced by TR and constructed a competitive endogenous RNA network for TR-induced right ventricular myocardial disease by integrating the interaction of lncRNA-miRNA-mRNA. In addition, we analyzed the immune infiltration using integrated data and the correlation of each immune-related gene with key genes of the integrated expression matrix. The present study identified 648 differentially expressed mRNAs, 201 differentially expressed miRNAs, and 163 differentially expressed lncRNAs. Protein-protein interaction network analysis confirmed that ADRA1A, AVPR1B, OPN4, IL-1B, IL-1A, CXCL4, ADCY2, CXCL12, GNB4, CCL20, CXCL8, and CXCL1 were hub genes. CTD-2314B22.3, hsa-miR-653-5p, and KIF17ceRNA; SRGAP3-AS2, hsa-miR-539-5p, and SHANK1; CERS6-AS1, hsa-miR-497-5p, and OPN4; INTS6-AS1, hsa-miR-4262, and NEURL1B; TTN-AS1, hsa-miR-376b-3p, and TRPM5; and DLX6-AS1, hsa-miR-346, and BIRC7 axes were obtained by constructing the ceRNA networks. Through the immune infiltration analysis, we found that the proportion of CD4 and CD8 T cells was about 20%, and the proportion of fibroblasts and endothelial cells was high. Our findings provide some insights into the mechanisms of RNA interaction in TR-induced right ventricular cardiomyopathy and suggest that lncRNAs are a potential therapeutic target for treating right ventricular myocardial disease induced by TR.
Keywords: immune cell infiltration; lncrna-mRNA co-expression network; long non-coding RNAs; right ventricular cardiomyopathy; tricuspid regurgitation.
Copyright © 2021 Tian, Yang, Ke, Yang, Zhong, Wang and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

