Offspring production of ovarian organoids derived from spermatogonial stem cells by defined factors with chromatin reorganization
- PMID: 34603780
- PMCID: PMC8463929
- DOI: 10.1016/j.jare.2021.03.006
Offspring production of ovarian organoids derived from spermatogonial stem cells by defined factors with chromatin reorganization
Abstract
Introduction: Fate determination of germline stem cells remains poorly understood at the chromatin structure level.
Objectives: Our research hopes to develop successful offspring production of ovarian organoids derived from spermatogonial stem cells (SSCs) by defined factors.
Methods: The offspring production from oocytes transdifferentiated from mouse SSCs with tracking of transplanted SSCs in vivo, single cell whole exome sequencing, and in 3D cell culture reconstitution of the process of oogenesis derived from SSCs. The defined factors were screened with ovarian organoids. We uncovered extensive chromatin reorganization during SSC conversion into induced germline stem cells (iGSCs) using high throughput chromosome conformation.
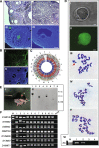
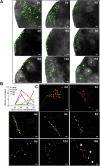
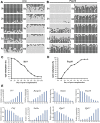
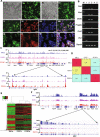
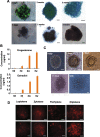
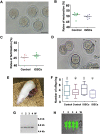
Results: We demonstrate successful production of offspring from oocytes transdifferentiated from mouse spermatogonial stem cells (SSCs). Furthermore, we demonstrate direct induction of germline stem cells (iGSCs) differentiated into functional oocytes by transduction of H19, Stella, and Zfp57 and inactivation of Plzf in SSCs after screening with ovarian organoids. We uncovered extensive chromatin reorganization during SSC conversion into iGSCs, which was highly similar to female germline stem cells. We observed that although topologically associating domains were stable during SSC conversion, chromatin interactions changed in a striking manner, altering 35% of inactive and active chromosomal compartments throughout the genome.
Conclusion: We demonstrate successful offspring production of ovarian organoids derived from SSCs by defined factors with chromatin reorganization. These findings have important implications in various areas including mammalian gametogenesis, genetic and epigenetic reprogramming, biotechnology, and medicine.
Keywords: 3D cell culture; Chromatin reorganization; Defined factors; Induced germline stem cells; Offspring production; Ovarian organoids.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Conflict of interest statement
The authors declared that there is no conflict of interest.
Figures


References
-
- Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y. Cell. 2017;170:367–381 e320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

