Diagnosis and prognosis models for hepatocellular carcinoma patient's management based on tumor mutation burden
- PMID: 34603786
- PMCID: PMC8463909
- DOI: 10.1016/j.jare.2021.01.018
Diagnosis and prognosis models for hepatocellular carcinoma patient's management based on tumor mutation burden
Abstract
Introduction: The development and prognosis of HCC involve complex molecular mechanisms, which affect the effectiveness of its treatment strategies. Tumor mutational burden (TMB) is related to the efficacy of immunotherapy, but the prognostic role of TMB-related genes in HCC has not yet been determined clearly.
Objectives: In this study, we identified TMB-specific genes with good prognostic value to build diagnostic and prognostic models and provide guidance for the treatment of HCC patients.
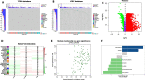
Methods: Weighted gene co-expression network analysis (WGCNA) was applied to identify the TMB-specific genes. And LASSO method and Cox regression were used in establishing the prognostic model.
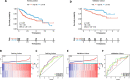
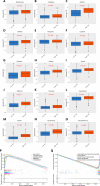
Results: The prognostic model based on SMG5 and MRPL9 showed patients with higher prognostic risk had a remarkedly poorer survival probability than their counterparts with lower prognostic risk in both a TCGA cohort (P < 0.001, HR = 1.93) and an ICGC cohort (P < 0.001, HR = 3.58). In addition, higher infiltrating fractions of memory B cells, M0 macrophages, neutrophils, activated memory CD4 + T cells, follicular helper T cells and regulatory T cells and higher expression of B7H3, CTLA4, PD1, and TIM3 were present in the high-risk group than in the low-risk group (P < 0.05). Patients with high prognostic risk had higher resistance to some chemotherapy and targeted drugs, such as methotrexate, vinblastine and erlotinib, than those with low prognostic risk (P < 0.05). And a diagnostic model considering two genes was able to accurately distinguish patients with HCC from normal samples and those with dysplastic nodules. In addition, knockdown of SMG5 and MRPL9 was determined to significantly inhibit cell proliferation and migration in HCC.
Conclusion: Our study helps to select patients suitable for chemotherapy, targeted drugs and immunotherapy and provide new ideas for developing treatment strategies to improve disease outcomes in HCC patients.
Keywords: Diagnosis; Hepatocellular carcinoma (HCC); Immune checkpoint; Prognosis; TMB.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2018;391:1301–1314. - PubMed
-
- Aravalli R.N., Steer C.J., Cressman E.N. Molecular mechanisms of hepatocellular carcinoma. Hepatology (Baltimore, MD) 2008;48:2047–2063. - PubMed
-
- Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. - PubMed
-
- Weston A.D., Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

