A Novel Seven Gene Signature-Based Prognostic Model to Predict Distant Metastasis of Lymph Node-Negative Triple-Negative Breast Cancer
- PMID: 34604089
- PMCID: PMC8481824
- DOI: 10.3389/fonc.2021.746763
A Novel Seven Gene Signature-Based Prognostic Model to Predict Distant Metastasis of Lymph Node-Negative Triple-Negative Breast Cancer
Abstract
Background: The prognosis of lymph node-negative triple-negative breast cancer (TNBC) is still worse than that of other subtypes despite adjuvant chemotherapy. Reliable prognostic biomarkers are required to identify lymph node-negative TNBC patients at a high risk of distant metastasis and optimize individual treatment.
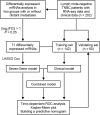
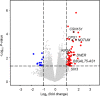
Methods: We analyzed the RNA sequencing data of primary tumor tissue and the clinicopathological data of 202 lymph node-negative TNBC patients. The cohort was randomly divided into training and validation sets. Least absolute shrinkage and selection operator Cox regression and multivariate Cox regression were used to construct the prognostic model.
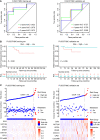
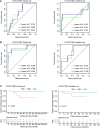
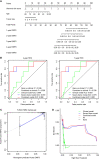
Results: A clinical prognostic model, seven-gene signature, and combined model were constructed using the training set and validated using the validation set. The seven-gene signature was established based on the genomic variables associated with distant metastasis after shrinkage correction. The difference in the risk of distant metastasis between the low- and high-risk groups was statistically significant using the seven-gene signature (training set: P < 0.001; validation set: P = 0.039). The combined model showed significance in the training set (P < 0.001) and trended toward significance in the validation set (P = 0.071). The seven-gene signature showed improved prognostic accuracy relative to the clinical signature in the training data (AUC value of 4-year ROC, 0.879 vs. 0.699, P = 0.046). Moreover, the composite clinical and gene signature also showed improved prognostic accuracy relative to the clinical signature (AUC value of 4-year ROC: 0.888 vs. 0.699, P = 0.029; AUC value of 5-year ROC: 0.882 vs. 0.693, P = 0.038). A nomogram model was constructed with the seven-gene signature, patient age, and tumor size.
Conclusions: The proposed signature may improve the risk stratification of lymph node-negative TNBC patients. High-risk lymph node-negative TNBC patients may benefit from treatment escalation.
Keywords: distant metastasis; modeling; prognostic biomarker; transcriptomics; triple-negative breast cancer.
Copyright © 2021 Peng, Lin, Jing, Su, Jin, Di and Shao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Construction and Validation of a Prognostic Risk Model for Triple-Negative Breast Cancer Based on Autophagy-Related Genes.Front Oncol. 2022 Feb 4;12:829045. doi: 10.3389/fonc.2022.829045. eCollection 2022. Front Oncol. 2022. PMID: 35186763 Free PMC article.
-
Integrative 3' Untranslated Region-Based Model to Identify Patients with Low Risk of Axillary Lymph Node Metastasis in Operable Triple-Negative Breast Cancer.Oncologist. 2019 Jan;24(1):22-30. doi: 10.1634/theoncologist.2017-0609. Epub 2018 Aug 6. Oncologist. 2019. PMID: 30082491 Free PMC article.
-
A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status.PLoS One. 2014 Jun 19;9(6):e100664. doi: 10.1371/journal.pone.0100664. eCollection 2014. PLoS One. 2014. PMID: 24945253 Free PMC article.
-
A combined hypoxia and immune gene signature for predicting survival and risk stratification in triple-negative breast cancer.Aging (Albany NY). 2021 Aug 2;13(15):19486-19509. doi: 10.18632/aging.203360. Epub 2021 Aug 2. Aging (Albany NY). 2021. PMID: 34341184 Free PMC article.
-
Advancement of prognostic models in breast cancer: a narrative review.Gland Surg. 2021 Sep;10(9):2815-2831. doi: 10.21037/gs-21-441. Gland Surg. 2021. PMID: 34733730 Free PMC article. Review.
Cited by
-
Identification of Prognostic Biomarkers for Breast Cancer Metastasis Using Penalized Additive Hazards Regression Model.Cancer Inform. 2023 Mar 21;22:11769351231157942. doi: 10.1177/11769351231157942. eCollection 2023. Cancer Inform. 2023. PMID: 36968522 Free PMC article.
-
Identification and panoramic analysis of drug response-related genes in triple negative breast cancer using as an example NVP-BEZ235.Sci Rep. 2023 Apr 12;13(1):5984. doi: 10.1038/s41598-023-32757-4. Sci Rep. 2023. PMID: 37045929 Free PMC article.
-
MLSP: A bioinformatics tool for predicting molecular subtypes and prognosis in patients with breast cancer.Comput Struct Biotechnol J. 2022 Nov 11;20:6412-6426. doi: 10.1016/j.csbj.2022.11.017. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467575 Free PMC article.
-
A comprehensive genomic and transcriptomic dataset of triple-negative breast cancers.Sci Data. 2022 Sep 24;9(1):587. doi: 10.1038/s41597-022-01681-z. Sci Data. 2022. PMID: 36153392 Free PMC article.
References
LinkOut - more resources
Full Text Sources

