MiR-1224 Acts as a Prognostic Biomarker and Inhibits the Progression of Gastric Cancer by Targeting SATB1
- PMID: 34604093
- PMCID: PMC8484804
- DOI: 10.3389/fonc.2021.748896
MiR-1224 Acts as a Prognostic Biomarker and Inhibits the Progression of Gastric Cancer by Targeting SATB1
Abstract
Objective: MiR-1224 has been reported to exhibit abnormal expression in several tumors. However, the expressing pattern and roles of miR-1224 in gastric cancer (GC) remain unclear. Our current research aimed to explore the potential involvement of miR-1224 in the GC progression.
Materials and methods: The expression of miR-1224 was examined in tissue samples of 128 GC patients and cell lines by RT-PCR. Besides, the associations of miR-1224 expressions with clinicopathologic features and prognosis of GC patients were analyzed. Then, the possible influences of miR-1224 on cell proliferation and cell migration were determined. Afterward, the molecular target of miR-1224 was identified using bioinformatics assays and confirmed experimentally. Finally, RT-PCR and Western blot assays were performed to investigate the effect of the abnormal miR-1224 expression on the EMT and Wnt/β-catenin pathway.
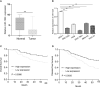
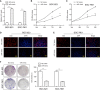
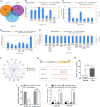
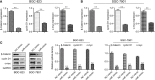
Results: miR-1224 was lowly expressed in the GC specimens and cell lines due to T classification and TNM stage. Survival assays demonstrated that GC patients with low expressions of miR-1224 possessed poor overall survivals. Moreover, in vitro and in vivo assays revealed that the overexpression of miR-1224 inhibited cell proliferation, migration, and invasion in GC cells. SATB homeobox 1 (SATB1) was verified as a direct target of miR-1224 in GC. Furthermore, β-catenin and c-myc were significantly inhibited in miR-1224-overexpression cells.
Conclusions: Our findings highlight the potential of miR-1224 as a therapeutic target and novel biomarker for GC patients.
Keywords: SATB1; Wnt/β-catenin; gastric cancer; metastasis; miR-1224; prognosis.
Copyright © 2021 Han, Sun, Hui, Tao, Liu and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway.Mol Cancer. 2018 Aug 22;17(1):126. doi: 10.1186/s12943-018-0874-1. Mol Cancer. 2018. PMID: 30134915 Free PMC article.
-
MiR-503 suppresses cell proliferation and invasion of gastric cancer by targeting HMGA2 and inactivating WNT signaling pathway.Cancer Cell Int. 2019 Jun 14;19:164. doi: 10.1186/s12935-019-0875-1. eCollection 2019. Cancer Cell Int. 2019. PMID: 31249473 Free PMC article.
-
LncRNA HOXA11-AS promotes migration and invasion through modulating miR-148a/WNT1/β-catenin pathway in gastric cancer.Neoplasma. 2020 May;67(3):492-500. doi: 10.4149/neo_2020_190722N653. Epub 2020 Feb 3. Neoplasma. 2020. PMID: 32009419
-
MiRNA-27a promotes the proliferation and invasion of human gastric cancer MGC803 cells by targeting SFRP1 via Wnt/β-catenin signaling pathway.Am J Cancer Res. 2017 Mar 1;7(3):405-416. eCollection 2017. Am J Cancer Res. 2017. PMID: 28401000 Free PMC article.
-
MiR-675 is frequently overexpressed in gastric cancer and enhances cell proliferation and invasion via targeting a potent anti-tumor gene PITX1.Cell Signal. 2019 Oct;62:109352. doi: 10.1016/j.cellsig.2019.109352. Epub 2019 Jun 28. Cell Signal. 2019. PMID: 31260797
Cited by
-
The Role and Mechanism of microRNA-1224 in Human Cancer.Front Oncol. 2022 Apr 14;12:858892. doi: 10.3389/fonc.2022.858892. eCollection 2022. Front Oncol. 2022. PMID: 35494023 Free PMC article. Review.
-
Regulation of the migration of colorectal cancer stem cells via the TLR4/MyD88 signaling pathway by the novel surface marker CD14 following LPS stimulation.Oncol Lett. 2023 Dec 18;27(2):60. doi: 10.3892/ol.2023.14194. eCollection 2024 Feb. Oncol Lett. 2023. PMID: 38192670 Free PMC article.
-
Identification of miRNA-mRNA regulatory network associated with the glutamatergic system in post-traumatic epilepsy rats.Front Neurol. 2022 Dec 23;13:1102672. doi: 10.3389/fneur.2022.1102672. eCollection 2022. Front Neurol. 2022. PMID: 36619916 Free PMC article.
-
SATB1 in cancer progression and metastasis: mechanisms and therapeutic potential.Front Oncol. 2025 Feb 25;15:1535929. doi: 10.3389/fonc.2025.1535929. eCollection 2025. Front Oncol. 2025. PMID: 40071088 Free PMC article. Review.
-
Joint global and local interpretation method for CIN status classification in breast cancer.Heliyon. 2024 Feb 28;10(5):e27054. doi: 10.1016/j.heliyon.2024.e27054. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38562500 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

