Taxonomic and Functional Dysregulation in Salivary Microbiomes During Oral Carcinogenesis
- PMID: 34604102
- PMCID: PMC8482814
- DOI: 10.3389/fcimb.2021.663068
Taxonomic and Functional Dysregulation in Salivary Microbiomes During Oral Carcinogenesis
Abstract
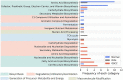
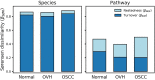
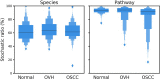
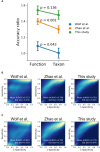
Exploring microbial community compositions in humans with healthy versus diseased states is crucial to understand the microbe-host interplay associated with the disease progression. Although the relationship between oral cancer and microbiome was previously established, it remained controversial, and yet the ecological characteristics and their responses to oral carcinogenesis have not been well studied. Here, using the bacterial 16S rRNA gene amplicon sequencing along with the in silico function analysis by PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2), we systematically characterized the compositions and the ecological drivers of saliva microbiome in the cohorts of orally healthy, non-recurrent oral verrucous hyperplasia (a pre-cancer lesion), and oral verrucous hyperplasia-associated oral cancer at taxonomic and function levels, and compared them with the re-analysis of publicly available datasets. Diversity analyses showed that microbiome dysbiosis in saliva was significantly linked to oral health status. As oral health deteriorated, the number of core species declined, and metabolic pathways predicted by PICRUSt2 were dysregulated. Partitioned beta-diversity revealed an extremely high species turnover but low function turnover. Functional beta-diversity in saliva microbiome shifted from turnover to nestedness during oral carcinogenesis, which was not observed at taxonomic levels. Correspondingly, the quantitative analysis of stochasticity ratios showed that drivers of microbial composition and functional gene content of saliva microbiomes were primarily governed by the stochastic processes, yet the driver of functional gene content shifted toward deterministic processes as oral cancer developed. Re-analysis of publicly accessible datasets supported not only the distinctive family taxa of Veillonellaceae and Actinomycetaceae present in normal cohorts but also that Flavobacteriaceae and Peptostreptococcaceae as well as the dysregulated metabolic pathways of nucleotides, amino acids, fatty acids, and cell structure were related to oral cancer. Using predicted functional profiles to elucidate the correlations to the oral health status shows superior performance than using taxonomic data among different studies. These findings advance our understanding of the oral ecosystem in relation to oral carcinogenesis and provide a new direction to the development of microbiome-based tools to study the interplay of the oral microbiome, metabolites, and host health.
Keywords: machine learning; microbiome dysbiosis; oral cancer; oral verrucous hyperplasia; saliva.
Copyright © 2021 Chen, Wu, Chiang, Chen, Wu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Al-Hebshi N. N., Borgnakke W. S., Johnson N. W. (2019). The Microbiome of Oral Squamous Cell Carcinomas: A Functional Perspective. Curr. Oral. Health Rep. 6, 145–160. doi: 10.1007/s40496-019-0215-5 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

