The Off-Targets of Clustered Regularly Interspaced Short Palindromic Repeats Gene Editing
- PMID: 34604217
- PMCID: PMC8484971
- DOI: 10.3389/fcell.2021.718466
The Off-Targets of Clustered Regularly Interspaced Short Palindromic Repeats Gene Editing
Abstract
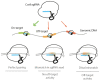
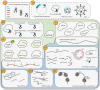
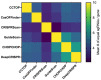
The repurposing of the CRISPR/Cas bacterial defense system against bacteriophages as simple and flexible molecular tools has revolutionized the field of gene editing. These tools are now widely used in basic research and clinical trials involving human somatic cells. However, a global moratorium on all clinical uses of human germline editing has been proposed because the technology still lacks the required efficacy and safety. Here we focus on the approaches developed since 2013 to decrease the frequency of unwanted mutations (the off-targets) during CRISPR-based gene editing.
Keywords: CRISPR/Cas9; Cas9 variants; DNA repair; gene editing; off-targets.
Copyright © 2021 Vicente, Chaves-Ferreira, Jorge, Proença and Barreto.
Conflict of interest statement
VMB runs a CRISPR-based gene editing service at Nova Medical School, a public university. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
[Advances in application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 system in stem cells research].Zhonghua Shao Shang Za Zhi. 2018 Apr 20;34(4):253-256. doi: 10.3760/cma.j.issn.1009-2587.2018.04.013. Zhonghua Shao Shang Za Zhi. 2018. PMID: 29690746 Review. Chinese.
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
Advances in therapeutic CRISPR/Cas9 genome editing.Transl Res. 2016 Feb;168:15-21. doi: 10.1016/j.trsl.2015.09.008. Epub 2015 Sep 26. Transl Res. 2016. PMID: 26470680 Review.
-
CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing.Trends Plant Sci. 2019 Dec;24(12):1102-1125. doi: 10.1016/j.tplants.2019.09.006. Epub 2019 Nov 11. Trends Plant Sci. 2019. PMID: 31727474 Review.
-
CRISPR/Cas gene therapy.J Cell Physiol. 2021 Apr;236(4):2459-2481. doi: 10.1002/jcp.30064. Epub 2020 Sep 22. J Cell Physiol. 2021. PMID: 32959897 Review.
Cited by
-
Advances and applications of CRISPR/Cas-mediated interference in Escherichia coli.Eng Microbiol. 2023 Nov 2;4(1):100123. doi: 10.1016/j.engmic.2023.100123. eCollection 2024 Mar. Eng Microbiol. 2023. PMID: 39628789 Free PMC article. Review.
-
Small Molecules for Enhancing the Precision and Safety of Genome Editing.Molecules. 2022 Sep 23;27(19):6266. doi: 10.3390/molecules27196266. Molecules. 2022. PMID: 36234804 Free PMC article. Review.
-
Macrophages derived from pluripotent stem cells: prospective applications and research gaps.Cell Biosci. 2022 Jun 20;12(1):96. doi: 10.1186/s13578-022-00824-4. Cell Biosci. 2022. PMID: 35725499 Free PMC article. Review.
-
Fast-Track and Integration-Free Method of Genome Editing by CRISPR/Cas9 in Murine Pluripotent Stem Cells.Front Cell Dev Biol. 2022 Mar 18;10:819906. doi: 10.3389/fcell.2022.819906. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35372368 Free PMC article.
-
Current concepts of the crosstalk between lncRNA and E2F1: shedding light on the cancer therapy.Front Pharmacol. 2024 Jul 25;15:1432490. doi: 10.3389/fphar.2024.1432490. eCollection 2024. Front Pharmacol. 2024. PMID: 39119602 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

