The Big Picture of Glioblastoma Malignancy: A Meta-Analysis of Glioblastoma Proteomics to Identify Altered Biological Pathways
- PMID: 34604635
- PMCID: PMC8482494
- DOI: 10.1021/acsomega.1c02991
The Big Picture of Glioblastoma Malignancy: A Meta-Analysis of Glioblastoma Proteomics to Identify Altered Biological Pathways
Abstract
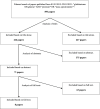
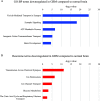
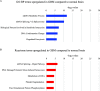
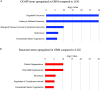
Glioblastoma is a highly malignant cancer with no effective treatment. It is vital to elucidate the mechanisms which drive glioblastoma in order to identify therapeutic targets. The differences in protein expression between glioblastoma, grade I-III glioma, and normal brain tissue reflect the functional alterations driving malignancy. However, proteomic analysis of glioblastoma has been hampered by the heterogeneity of glioblastoma and the variety of methodology used in its study. To reduce these inconsistencies, we performed a meta-analysis of the literature published since 2015, including 14 datasets from eight papers comparing the whole proteome of glioblastoma to normal brain or grade I-III glioma. We found that 154 proteins were commonly upregulated and 116 proteins were commonly downregulated in glioblastoma compared to normal brain. Meanwhile, 240 proteins were commonly upregulated and 125 proteins were commonly downregulated in glioblastoma compared to grade I-III glioma. Functional enrichment analysis revealed upregulation of proteins involved in mRNA splicing and the immune system and downregulation of proteins involved in synaptic signaling and glucose and glutamine metabolism. The identification of these altered biological pathways provides a basis for deeper investigation in the pursuit of an effective treatment for glioblastoma.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Verhaak R. G. W.; Hoadley K. A.; Purdom E.; Wang V.; Qi Y.; Wilkerson M. D.; Miller C. R.; Ding L.; Golub T.; Mesirov J. P.; Alexe G.; Lawrence M.; O’Kelly M.; Tamayo P.; Weir B. A.; Gabriel S.; Winckler W.; Gupta S.; Jakkula L.; Feiler H. S.; Hodgson J. G.; James C. D.; Sarkaria J. N.; Brennan C.; Kahn A.; Spellman P. T.; Wilson R. K.; Speed T. P.; Gray J. W.; Meyerson M.; Getz G.; Perou C. M.; Hayes D. N. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. 10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
-
- Wang Q.; Hu B.; Hu X.; Kim H.; Squatrito M.; Scarpace L.; deCarvalho A. C.; Lyu S.; Li P.; Li Y.; Barthel F.; Cho H. J.; Lin Y.-H.; Satani N.; Martinez-Ledesma E.; Zheng S.; Chang E.; Sauvé C.-E. G.; Olar A.; Lan Z. D.; Finocchiaro G.; Phillips J. J.; Berger M. S.; Gabrusiewicz K. R.; Wang G.; Eskilsson E.; Hu J.; Mikkelsen T.; DePinho R. A.; Muller F.; Heimberger A. B.; Sulman E. P.; Nam D.-H.; Verhaak R. G. W. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. 10.1016/j.ccell.2017.06.003. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

