A natural language processing pipeline to synthesize patient-generated notes toward improving remote care and chronic disease management: a cystic fibrosis case study
- PMID: 34604710
- PMCID: PMC8480545
- DOI: 10.1093/jamiaopen/ooab084
A natural language processing pipeline to synthesize patient-generated notes toward improving remote care and chronic disease management: a cystic fibrosis case study
Abstract
Objectives: Patient-generated health data (PGHD) are important for tracking and monitoring out of clinic health events and supporting shared clinical decisions. Unstructured text as PGHD (eg, medical diary notes and transcriptions) may encapsulate rich information through narratives which can be critical to better understand a patient's condition. We propose a natural language processing (NLP) supported data synthesis pipeline for unstructured PGHD, focusing on children with special healthcare needs (CSHCN), and demonstrate it with a case study on cystic fibrosis (CF).
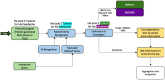
Materials and methods: The proposed unstructured data synthesis and information extraction pipeline extract a broad range of health information by combining rule-based approaches with pretrained deep-learning models. Particularly, we build upon the scispaCy biomedical model suite, leveraging its named entity recognition capabilities to identify and link clinically relevant entities to established ontologies such as Systematized Nomenclature of Medicine (SNOMED) and RXNORM. We then use scispaCy's syntax (grammar) parsing tools to retrieve phrases associated with the entities in medication, dose, therapies, symptoms, bowel movements, and nutrition ontological categories. The pipeline is illustrated and tested with simulated CF patient notes.
Results: The proposed hybrid deep-learning rule-based approach can operate over a variety of natural language note types and allow customization for a given patient or cohort. Viable information was successfully extracted from simulated CF notes. This hybrid pipeline is robust to misspellings and varied word representations and can be tailored to accommodate the needs of a specific patient, cohort, or clinician.
Discussion: The NLP pipeline can extract predefined or ontology-based entities from free-text PGHD, aiming to facilitate remote care and improve chronic disease management. Our implementation makes use of open source models, allowing for this solution to be easily replicated and integrated in different health systems. Outside of the clinic, the use of the NLP pipeline may increase the amount of clinical data recorded by families of CSHCN and ease the process to identify health events from the notes. Similarly, care coordinators, nurses and clinicians would be able to track adherence with medications, identify symptoms, and effectively intervene to improve clinical care. Furthermore, visualization tools can be applied to digest the structured data produced by the pipeline in support of the decision-making process for a patient, caregiver, or provider.
Conclusion: Our study demonstrated that an NLP pipeline can be used to create an automated analysis and reporting mechanism for unstructured PGHD. Further studies are suggested with real-world data to assess pipeline performance and further implications.
Keywords: artificial intelligence; chronic disease; cystic fibrosis; natural language processing; patient notes.
© The Author(s) 2021. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures

Similar articles
-
Extracting Medical Information From Free-Text and Unstructured Patient-Generated Health Data Using Natural Language Processing Methods: Feasibility Study With Real-world Data.JMIR Form Res. 2023 Mar 7;7:e43014. doi: 10.2196/43014. JMIR Form Res. 2023. PMID: 36881467 Free PMC article.
-
Identification of Preanesthetic History Elements by a Natural Language Processing Engine.Anesth Analg. 2022 Dec 1;135(6):1162-1171. doi: 10.1213/ANE.0000000000006152. Epub 2022 Jul 15. Anesth Analg. 2022. PMID: 35841317 Free PMC article.
-
A Hybrid Model for Family History Information Identification and Relation Extraction: Development and Evaluation of an End-to-End Information Extraction System.JMIR Med Inform. 2021 Apr 22;9(4):e22797. doi: 10.2196/22797. JMIR Med Inform. 2021. PMID: 33885370 Free PMC article.
-
Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review.JMIR Med Inform. 2019 Apr 27;7(2):e12239. doi: 10.2196/12239. JMIR Med Inform. 2019. PMID: 31066697 Free PMC article. Review.
-
From admission to discharge: a systematic review of clinical natural language processing along the patient journey.BMC Med Inform Decis Mak. 2024 Aug 29;24(1):238. doi: 10.1186/s12911-024-02641-w. BMC Med Inform Decis Mak. 2024. PMID: 39210370 Free PMC article.
Cited by
-
Extracting Medical Information From Free-Text and Unstructured Patient-Generated Health Data Using Natural Language Processing Methods: Feasibility Study With Real-world Data.JMIR Form Res. 2023 Mar 7;7:e43014. doi: 10.2196/43014. JMIR Form Res. 2023. PMID: 36881467 Free PMC article.
-
Virtual monitoring in CF - the importance of continuous monitoring in a multi-organ chronic condition.Front Digit Health. 2023 May 4;5:1196442. doi: 10.3389/fdgth.2023.1196442. eCollection 2023. Front Digit Health. 2023. PMID: 37214343 Free PMC article. Review.
-
"Hey Siri, Help Me Take Care of My Child": A Feasibility Study With Caregivers of Children With Special Healthcare Needs Using Voice Interaction and Automatic Speech Recognition in Remote Care Management.Front Public Health. 2022 Mar 3;10:849322. doi: 10.3389/fpubh.2022.849322. eCollection 2022. Front Public Health. 2022. PMID: 35309210 Free PMC article.
-
Using electronic health records for clinical pharmacology research: Challenges and considerations.Clin Transl Sci. 2024 Jul;17(7):e13871. doi: 10.1111/cts.13871. Clin Transl Sci. 2024. PMID: 38943244 Free PMC article. Review.
-
Artificial intelligence in the care of children and adolescents with chronic diseases: a systematic review.Eur J Pediatr. 2024 Dec 14;184(1):83. doi: 10.1007/s00431-024-05846-3. Eur J Pediatr. 2024. PMID: 39672974 Free PMC article.
References
-
- McPherson M, Arango P, Fox H, et al.A new definition of children with special health care needs. Pediatrics 1998; 102 (1 Pt 1): 137–40. - PubMed
-
- 2009. –C. The National Survey of Children with Special Health Care Needs. https://mchb.hrsa.gov/sites/default/files/mchb/Data/NSCH/nscshcn0910-cha... Accessed June 9, 2021.
LinkOut - more resources
Full Text Sources
